Abstract
Simple retroviruses induce tumors by integrating into the host genome, activating cellular oncogenes and microRNAs, or inactivating tumor suppressor genes. The identification of these genes elucidates molecular mechanisms of tumorigenesis. In this study, we identified avian leukosis virus (ALV) proviral integration sites in rapid-onset B cell lymphomas arising <12 weeks after infection of chicken embryos. By using inverse PCR, 28 unique viral integration sites were identified in rapid-onset tumors. Integrations in the telomerase reverse transcriptase (TERT) promoter/enhancer region were observed in four different tumors, suggesting that this is a common integration site. These provirus integrations ranged from 217 to 2,584 bp upstream of the TERT transcription initiation site and were all in the opposite transcriptional orientation to TERT. Southern blots of tumor samples demonstrated that these integrations are clonal and therefore occurred early in the process of tumorigenesis. Real-time RT-PCR showed overexpression of TERT mRNA in tumors harboring viral integrations in the TERT promoter. Telomerase activity was also up-regulated in these tumors; however, telomere-length alterations were not detected. Furthermore, viral LTR sequences directly enhanced the expression of luciferase reporters containing the TERT promoter sequences. This study documents retroviral up-regulation of cellular TERT by insertional activation to initiate or enhance tumor progression.
Keywords: retroviral tagging, chicken tumors, chicken telomerase
Cancer development is a multistep process requiring sequential changes in various genes and cellular pathways (1). The critical role of telomerase in tumorigenesis is evident by its up-regulation in >90% of human cancers, especially malignant, late-stage tumors (2). The classic pathway of oncogenesis in somatic cells is thought to be triggered by shortened, dysfunctional telomeres (3), which lead to significant chromosome instability and fusion-bridge events (4). Eventually, some of these cells extend their telomeres either by reactivation of telomerase activity or by the alternative lengthening of telomeres (ALT) mechanism (3). This restores genomic stability, bypasses cell cycle arrest and establishes a cancer progenitor cell. In contrast, telomerase reactivation is not necessary in cancers originating from stem cells due to their persistent high telomerase activity (5).
Retroviruses provide a unique method of studying cancer progression, because they can induce tumors in animal hosts by insertionally activating or, more rarely, inactivating cellular genes. When retroviruses integrate into the host genome, they frequently place a host gene under the control of viral regulatory elements (6). This mimics natural cancer development in humans because it selects for a complement of mutations that support the cancer phenotype. Thus, retrovirus-tagged oncogene discovery has been a potent tool in revealing potential oncogenes and tumor-suppressor genes in both chicken and mouse model systems (6, 7). To classify a virus-tagged gene as a potential cancer gene, it must be present in multiple independent tumors. Because viral integration is random, common integration sites are believed to result from strong biological selection (7).
Avian leukosis virus (ALV) is a simple retrovirus that does not encode an oncogene but that can induce tumors by integrating near oncogenes, introducing a strong promoter or enhancer sequence (8–12). Typically, ALV induces B cell lymphomas to develop in ≈6 months and frequently involves proviral integrations, resulting in deregulation of the cellular transcription factor, myc, as well as bic (precursor to microRNA 155) (10–12). In addition, B cell lymphomas with a more rapid onset (lethal in <3 months) have been observed with clonal proviral insertions into the myb gene locus, resulting in overexpression of a truncated Myb transcription factor (8, 9). Because the cancer phenotype appears to involve a complement of cellular abnormalities, it is probable that additional oncogenes are altered in these tumors.
When 10-day-old chicken embryos were infected with certain mutant strains of ALV, B cell lymphomas caused death in up to 75% of the chickens by 10 weeks after hatching (13). We performed inverse PCR on genomic DNA from these lymphomas and identified 28 unique viral integration sites in tumors from eight different birds. Integrations into the telomerase reverse transcriptase (TERT) gene were independently identified in tumors in four different birds, making TERT a common integration site. The integrations all occurred within the TERT promoter/enhancer region, 0.2–2.6 kb upstream of the TERT transcription initiation site, and all were in the opposite transcriptional orientation. Because human TERT is only thought to be expressed in stem cells and cancer cells (5), our finding of a common insertion site in the chicken TERT gene appeared to be relevant to tumorigenesis.
Furthermore, real-time RT-PCR showed up-regulation of TERT mRNA in all tumors harboring ALV integrations upstream of the TERT gene. This up-regulation was due to the regulatory effects of the inserted viral enhancer. Furthermore, functional telomerase activity was found to be increased when measured by the telomeric repeat amplification protocol (TRAP). Because these tumors were clonal for viral integration into TERT, our data suggest that up-regulation of TERT is an early event in malignancy. Interestingly, telomerase seems to have exerted its oncogenic effects independent of changes in telomere length.
Results
Proviral Integration into myb Is Not Always Clonal in Rapid-Onset Avian Lymphomas.
ALV induces B cell lymphomas associated with clonal proviral integrations into various proto-oncogenes, including myc and myb (8–12, 14). To determine whether myb integration acts as an initiating event in tumorigenesis in these chickens, as it does with previously studied rapid-onset ALV-induced lymphomas (8, 9, 14), we examined the clonality of myb integrations in previously studied tumors (13). If the tumor is clonal for myb, we expect to see equal signal from both the rearranged chromosome and the WT chromosome. Because PCR analysis had shown that all of these tumors had integrations in the myb locus (15), we were surprised to find that only 2 of 10 tumors analyzed by Southern blotting (A4 and C3) had clonal integrations in myb (data not shown). This finding suggests that myb integrations may not have been an early event in the formation of all of these tumors, and proviral integrations into other genes might have a more significant contribution to initiating tumor formation.
Most Proviral Insertion Sites in Tumors Are Near or Within Genes.
To identify additional genes involved in the induction of these tumors, we selected tumors from eight different chickens for further analysis. Inverse PCR was used to identify proviral integration sites in genomic tumor DNA, usually obtained from the liver, where the B cell lymphomas had metastasized. Twenty-eight unique proviral insertion sites were isolated from these eight tumors [supporting information (SI) Table 1]; this is almost certainly an underestimate of the total number of integration sites in these tumors. Nineteen of these integrations were mapped within gene transcripts, and an additional seven were mapped within 4 kb of transcribed cellular genes. Only 2 of these 28 integrations were intergenic, suggesting a strong selection in the tumors for integration sites in or near genes. Integration sites were found on chromosomes 1, 2, 3, 4, 7, 8, 9, and 17. Of these 26 genic integration sites, 1 was exonic and 18 were intronic. Six occurred upstream (0.4–3.9 kb) in the enhancer/promoter region of genes, and one was 3.4 kb downstream of a gene. Thirteen integrations were found in the same transcriptional orientation as the nearest gene, whereas the others were antisense, suggesting that the orientation of integration was random. Some of the integration sites identified were in or near previously identified proto-oncogenes, including myb, vav-3, whsc1, and tpd52.
The TERT Promoter/Enhancer Region Is a Common ALV Integration Site.
Of all of the viral integration sites identified by inverse PCR to date, the only integration site common to multiple tumors was upstream of the TERT gene. This integration locus was identified in four different tumors from different birds, termed C2, C6, C7, and D2 (Fig. 1A and SI Table 1). The integration site of tumor C6 was found on chromosome 2 at nucleotide 17855498 of Gallus gallus assembly 2.1, which is 2,584 bp upstream of the TERT transcription start site. Similarly, the integration site of tumor C7 was detected by inverse PCR at a position 1,088 bp upstream of TERT. On the basis of direct PCR, we found evidence of integration sites 217 and 413 bp upstream of TERT in tumors C7 and D2, respectively. Remarkably, all four proviral integrations were clustered within a 2.4-kb stretch in the promoter/enhancer region of TERT, and all four proviruses were in the reverse transcriptional orientation relative to TERT. This finding is suggestive of a strong selective preference for this common integration site location and orientation in these four tumors.
Fig. 1.
Proviral integration sites in the TERT and myb gene. (A) Schematic view of proviral integration sites and predicted transcription-factor-binding sites in the TERT. The position of identified proviral integration sites in the chTERT promoter/enhancer region are depicted as the number of nucleotides upstream of the TERT transcriptional initiation site. The transcription-factor-binding sites were predicted by Transcription Element Search Software (TESS). The arrows indicate the EcoRI sites, and the black bar shows the location of the probe used in Southern blot analysis. (B) Southern blot analysis was carried out with EcoRI-digested genomic DNA from normal uninfected liver (NL) and bursa (NB), liver tumors, or bursa tumors by using a probe in TERT exon 2 (nt 1,179–2,105). (C) The blot was stripped and reprobed with a probe spanning from myb intron 3 to exon 5. The integrations of proviral sequences introduce additional EcoRI sites and rearrange the WT 5.3-kb alleles to a 4.0-kb band.
To confirm the inverse PCR results, we used a viral LTR-specific primer, together with a TERT exon 1-specific primer, to directly amplify the DNA sequences flanking the viral integration site. The amplified bands were purified, cloned and sequenced revealing that the viral/cellular junctions matched the inverse PCR results for tumors C6 and C7 (data not shown). In addition, in tumor C7, we observed evidence for multiple integration sites upstream of TERT by direct PCR, suggesting that this tumor may be oligoclonal.
To further confirm the provirus integration sites in these tumors and to ask what fraction of the cellular genes were rearranged, the tumor genomic DNA was digested with EcoR1 and a Southern blot analysis was performed using a probe complementary to TERT exon 2 (nt +1179–2105). In the Southern blot analysis shown in Fig. 1B, the normal TERT gene (NB and NL) exhibited a 6.7-kb band, as predicted from the EcoR1 restriction sites at −4,489 bp and +2,223 bp in the TERT gene (Fig. 1A). Because the viral LTRs, gag, pol, and env genes contain EcoRI sites, the integration of intact proviral DNA would introduce five additional EcoRI sites upstream of TERT, disrupting the normal 6.7-kb band and generating a smaller, rearranged band when probed with the TERT probe.
On the basis of the sites of integration mapped by inverse PCR and direct PCR, the sizes of the rearranged bands were predicted to be 4.8 kb for C6 and 2.6 kb for D2. The C7 tumor was predicted to generate two rearranged bands with sizes of 3.3 kb and 2.4 kb. These tumors all were found to have a normal TERT allele and a rearranged allele of the expected size (Fig. 1B). In contrast, the A4 tumor contained no proviral integration in TERT and only showed the 6.7-kb normal TERT allele. In addition, we observed a rearranged band of ≈3.0 kb for C2L. On the basis of the size of the rearranged band, we predicted that the proviral integration site for C2 occurred at a position ≈0.8 kb upstream of the TERT transcript. Inverse PCR had detected another integration site in Tumor C2 at position −2,323, suggesting multiple ALV integrations into TERT in the same tumor. Southern blotting and direct PCR also revealed multiple rearranged bands for the tumor C7. Because the rearranged bands for tumors C2 and C7 were weaker than the WT allele, we conclude tumors C2 and C7 are oligoclonal for integration into the TERT locus, whereas C6 and D2 are clonal. These results, together with the inverse PCR data and direct genomic PCR results, confirm that the upstream regulatory region of the TERT gene is a common provirus insertion site in short-latency B cell lymphomas. We also observed clonal TERT rearrangement in the C6 bursa (data not shown), suggesting that this event is likely to occur early in tumor induction.
To determine whether these same tumor samples also have clonal viral integrations in the myb locus, we stripped the membrane and blotted with a myb-specific probe, containing sequences from myb intron 3 to exon 5. Normal bursa and liver, and tumors (C2, C6, C7, D2) were found to contain only the WT myb allele (Fig. 1C). Because these tumors had been shown to contain myb integration by nested PCR (15), these data suggested that myb integration is not clonal in these tumors. In contrast, the equal signal from the rearranged and WT allele in tumor A4 (Fig. 1C) and C3 (data not shown) indicated clonal rearrangement of myb but not of TERT.
TERT mRNA Expression Was Elevated in Tumors with Proviral Integrations in TERT.
Up-regulation of TERT expression has been observed in >90% of human tumors (2), suggesting that its activation is required for tumor initiation and/or progression. Our finding of four common ALV integrations in the TERT promoter region in the antisense orientation led us to propose that these tumor cells used the viral LTR enhancer to promote elevated or deregulated TERT gene expression. To test this hypothesis, we first sought to characterize TERT mRNA expression levels in the tumors containing TERT proviral integrations. Using real-time RT-PCR with gene-specific primers, we compared three bursa tumors and eight metastasized liver tumors with and without viral integrations in TERT. The expression data in Figs. 2 and 3 were normalized either to GAPDH mRNA or 18S ribosomal RNA levels with similar results.
Fig. 2.
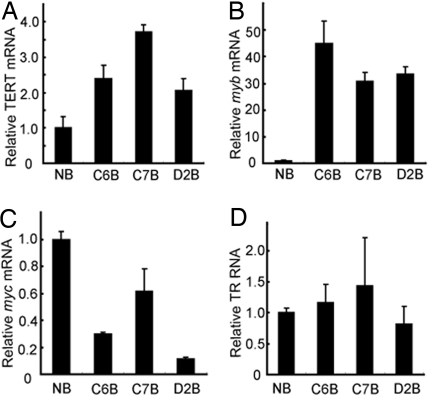
Expression levels of TERT (A), myb (B), myc (C) and TR (D) in normal bursa and bursa tumors. RNA levels were measured by real-time RT-PCR using gene-specific primers and were normalized relative to GAPDH mRNA levels. Bursa tumor RNA levels were plotted relative to levels in uninfected normal tissues.
Fig. 3.
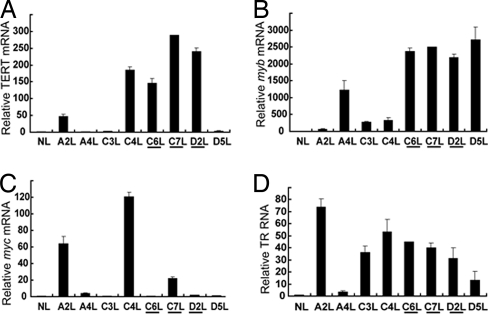
Expression levels of TERT (A), myb (B), myc (C) and TR (D) in normal liver and liver tumors. RNA levels were measured as in Fig. 2. The underlined samples are tumors with ALV integration sites in TERT.
We found that the tumors with TERT proviral integrations were all associated with elevated levels of TERT mRNA. TERT mRNA levels in bursa tumors C6, C7, and D2 increased 2.4-, 3.7-, and 2.1-fold, respectively, relative to levels in normal bursa (Fig. 2A). TERT mRNA levels in normal liver were 60-fold less than in normal bursa (data not shown), but in liver tumors, the TERT levels increased 145-, 288-, and 240-fold, respectively, relative to normal liver (Fig. 3A). Among the eight liver tumors analyzed by real-time RT-PCR, tumors A2 and C4 also had significant increases in TERT mRNA levels, 47- and 186-fold, respectively, over normal liver, even though we did not detect proviral integration into their TERT locus (Fig. 3A).
We evaluated the possibility that our B cell tumor samples obtained from the liver were contaminated with normal liver tissue by real-time RT PCR analysis, comparing expression of liver-specific albumin with B cell-specific IgG (SI Fig. 6). We discarded some tumor samples with high relative albumin expression. Most of the rest were >80% B cell specific, except for tumors C6 and C2, which appeared to be composed of ≈2/3 liver on the basis of our analysis. Three of the lymphomas with significant levels of B cells in the liver (SI Fig. 6), presumably due to tumor metastases, failed to show any TERT expression (A4, C3, D5), including two of the tumors with clonal integrations into myb (data not shown).
Because binding sequences for myb and myc are present in the TERT promoter/enhancer (Fig. 1A), this suggests a possible regulatory role for Myb and Myc in TERT gene expression. Thus, we next examined the mRNA levels of myb and myc in these tumors by real-time RT-PCR. Significantly, the myb mRNA levels in three bursal tumors with TERT viral integration, C6, C7, and D2, were 45-, 31-, and 33-fold higher than normal bursa levels (Fig. 2B). In the corresponding liver tumors, the myb expression levels were all >2,000-fold higher than normal liver myb levels (Fig. 3B). Normal bursa had 80-fold higher myb levels than normal liver. As expected, myb mRNA was also up-regulated in many tumors without TERT viral integration, including A4 and C3, which had ALV integrations in myb (data not shown).
The myc mRNA levels were not detectably up-regulated in the bursa tumors studied (Fig. 2C) but were shown to be 22 times higher than normal liver in the C7 liver tumor, which has a TERT viral integration (Fig. 3C). myc was also elevated 64- and 120-fold, respectively, in A2 and C4 liver tumors, which did not have known viral insertions in the TERT locus (Fig. 3C). Interestingly, A2 and C4 tumors had no known proviral integration in TERT but had elevated TERT mRNA expression, and they both exhibited up-regulation of myc mRNAs (Fig. 3C), suggesting that Myc may play a role in regulating TERT expression in these tumors. From this we conclude that proviral integrations in TERT are associated with up-regulation of TERT mRNA.
Analysis of TR RNA Levels in Bursa and Liver Tumors.
A functional telomerase heterodimer consists of both the enzymatic component TERT and the RNA template component TR (16). Although it has been suggested that TR is constitutively expressed in all tissues (17), recent studies have shown that TR levels are not elevated in normal tissues and only up-regulated in some tumors (18). To examine whether TR expression has been affected in the short-latency chicken tumors, we included TR-specific primers in the real-time RT-PCR studies. TR levels in all three (C6, C7, D2) bursa tumors analyzed were not found to be up-regulated in this assay (Fig. 2D). However, in the metastatic liver tumors, viral integration in TERT was associated with 45-, 40-, and 31-fold higher expression of TR compared with normal uninfected liver tissue (Fig. 3D). Of five tested tumors without viral integrations in TERT, four tumors, A2, C3, C4, and D5, also had increased TR RNA levels 74-, 36-, 53-, and 13-fold higher than normal liver. However, normal bursa was found to have 28-fold higher levels of TR than normal liver (data not shown).
Telomerase Activity Was Activated in Tumors with TERT Integrations.
Because TERT was up-regulated in these tumors, we next asked whether this led to an increase in telomerase activity. We used TRAP (19) to determine the telomerase enzyme activity in these tumors. Fig. 4 shows a typical TRAP gel autoradiogram. We quantified the signals from telomeric repeats and found that normal bursa carried significant levels of TRAP activity, confirming the existence of developmentally immature cells in this early bursa. Bursas taken from birds with tumors (C6, C7, and D2) having TERT integrations were associated with 2.0-, 3.23-, and 3.13-fold increases in TRAP activity respectively, when normalized to normal bursa. Normal liver and liver tumors (A2, A4, C3, and D5) without TERT integrations showed very low levels of telomerase activity. Conversely, liver tumors with TERT integrations (C6, C7, and D2) and a fourth tumor with high TERT mRNA levels (C4) were associated with 1.6-, 2.9-, 2.6-, and 2.0-fold increases in activity, respectively, when compared with normal bursa and were associated with 7.0-, 12.6-, 11.4-, and 8.7-fold increases, respectively, when compared with normal liver.
Fig. 4.
Viral integrations in TERT result in enhanced telomerase activity in tumors. (A) Telomerase activity in each tumor was measured by TRAP. The KBMC tumor cell line was used as a positive control. (B) Quantification of TRAP. All samples were normalized to normal bursa (NB). TRAP activity and the relative telomerase activities were plotted. The underlined samples are tumors with ALV integration sites in TERT.
To elucidate the potential role of proviral integration activating TERT in tumorigenesis, we examined the telomere length of tumors with or without TERT integration. Genomic DNA obtained from normal and tumor tissues were digested by HaeIII, a restriction enzyme that lacks digestion sites in the telomere. Southern blotting was carried out using a probe against the telomeric repeat sequence. There was no apparent change in telomere length among these samples (data not shown). This finding further supports our hypothesis that insertional activation of TERT by ALV was a primary event in tumorigenesis and not a later event triggered by unstable short telomere ends. Furthermore, this finding suggests TERT may have an alternate role in promoting tumor development.
ALV U3 LTR Sequence Enhanced Transcription of a Reporter Driven by the TERT Promoter.
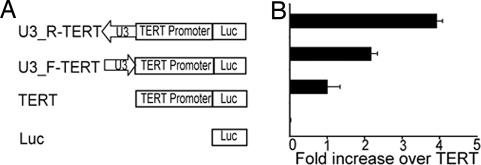
Because the four integrated proviruses were all upstream and oriented in the opposite direction relative to TERT transcription, we proposed that the increase in TERT mRNA levels was likely due to the enhancer elements in the viral LTR acting in cis to activate gene transcription, as has been seen in other tumors (6). To address this hypothesis, we fused U3 enhancer sequences from the viral LTR in the sense or antisense orientation 2.6 kb upstream of the TERT transcription initiation site (Fig. 5A). This position corresponds to the site of proviral insertion in tumor C6. We compared the transcriptional activity of the WT TERT promoter alone and the fused U3-TERT promoter in a luciferase reporter system in chicken embryo fibroblasts (CEFs). Fig. 5B shows that the WT TERT promoter/enhancer only very weakly enhances luciferase activity in CEFs, which is consistent with the fact that TERT is expressed at a very low level in somatic tissue. Insertion of the LTR in the sense orientation increased the luciferase levels ≈2-fold. Luciferase levels increased ≈4-fold when the LTR enhancer was inserted in the antisense orientation and compared with the WT chTERT promoter alone. This transcriptional enhancement appears to be attributable to an orientation-dependent activity of the ALV regulatory elements.
Fig. 5.
ALV enhancers increase expression of luciferase reporters containing the TERT promoter/enhancer region. (A) Schematic view of firefly luciferase reporter constructs bearing chTERT promoter/enhancer (2.6 kb upstream region) with or without LTR enhancer. (B) CEF cells were transiently transfected with firefly luciferase reporter and a control renilla luciferase construct. The expression levels of each firefly luciferase reporter was normalized by renilla luciferase and then plotted against the TERT reporter.
Similar results were observed after transfection of these constructs into a B cell tumor line, KBMC cells, which were derived by transformation with Rel (20) (data not shown). Interestingly, the expression levels of all these constructs in KBMC cells were 7-fold higher overall than those in CEFs. This finding suggests that additional transcription factors, such as Rel, active in the KBMC tumor cell line may also be contributing to TERT expression as has been proposed by Bose and coworkers (21). These results suggest that integration of the viral LTR in the chTERT promoter/enhancer region can directly enhance the transcription of TERT mRNA from its endogenous promoter. The observed enhancer effect also seems to be independent of cell type, yielding similar results in both CEF and KBMC cells.
Discussion
ALV infection of cultured chicken embryo fibroblasts, with no selection for tumor formation, resulted in nearly random integrations in the genome (approximately one-third of the insertions were associated with genes) in the work of Bushman and coworkers (22). Furthermore, endogenous ALV integrations in chickens were found to be predominantly intergenic, and this was attributed to a purifying coevolution of virus and host (22). In contrast, 93% of the ALV integration sites we identified in rapid-onset B cell lymphomas are within or very near genes, suggesting that selectivity of certain integration sites is occurring in tumor cells. Consistent with this finding, we observed integrations into several sites of known proto-oncogenes (Myb, Vav3, WHSC1, and TPD52), as well as four common integration sites in the TERT upstream region.
ALV proviral integrations in four different B cell lymphomas were in the TERT gene, varying from 0.2 to 2.6 kb upstream of the TERT transcription initiation site, making TERT a common insertion site for ALVs in B cell lymphomas. Remarkably, all five integrations occurred in the reverse orientation, suggesting that enhancers in the proviral LTR may cis-activate TERT transcription. Alternatively, proviral DNA insertion may have disrupted some TERT repressor-binding sequences. Finally, transcription factors acting in trans on TERT may have been induced by insertional mutagenesis (myc or rel, for example). We found that the viral LTR enhanced telomerase promoter activity, especially in the reverse orientation, and conclude that ALV LTR has a direct regulatory effect on TERT transcription.
Two human viruses, hepatitis B virus and papilloma virus, have also been observed to integrate into TERT in a small number of liver and cervical tumors, respectively (23, 24), suggesting proviral integration in TERT may be of more general importance in oncogenesis. However, the effect of viral integration into TERT on its expression was not determined in these previous studies. In addition, telomerase is transactivated by v-Rel during transformation of chicken lymphoid cells and fibroblasts (21). Furthermore, Marek's disease herpesvirus (MDV) encodes a viral TR that is 88% homologous to host TR and that contributes to induction of chicken T cell lymphomas (25).
In confirming the viral integrations by Southern blotting, we demonstrated that the proviral integrations upstream of TERT, in tumors C6 and D2, were clonal. Clonality suggests that the TERT integration exists in every cell in the tumor. Thus, the progenitor B cell leading to these tumors must have harbored a TERT integration very early in tumorigenesis. This may have provided a growth advantage to these cells, making them more susceptible to further selective mutations and malignant transformation. Conversely, myb integration sites in these four tumors were not observed by Southern blot analysis but only by direct PCR (15). This finding suggests that the myb integrations were not clonal and may represent a later event distinct from the clonal myb-lymphomas described in refs. 8, 9, and 26. Alternatively, myb may be needed for tumor initiation, but not its progression. Consistent with this, clonal proviral integrations into myb have been observed in the bursas of some of these birds (15). We hypothesize that tumorigenesis was triggered by activation of TERT by proviral integration in these four birds. Nevertheless, two other tumors in this group, A4 and C3, were clonal for myb integrations but showed no clonal integrations into TERT, suggesting that there are two different pathways involved in formation of these rapid-onset tumors.
After confirming the proviral integrations by Southern blot analysis, we sought to examine the viral enhancer's capacity to up-regulate TERT expression by real-time RT-PCR assays. In these experiments, TERT mRNA levels in tumor tissue were 2.1- to 3.7-fold higher than normal bursa and 150- to 300-fold higher than normal liver TERT mRNA levels. This large variation in gene expression enhancement is because, in 10-week-old chickens, the bursa possesses 60-fold higher levels of TERT mRNA than the liver (data not shown). This finding was not surprising because chicken bursa contains hematopoietic stem cells, which would be expected to express high levels of telomerase (27). In contrast, in the liver, hepatocyte progenitors are normally quiescent and only conditionally reactivated under circumstances of injury or need.
Based on the absence of telomerase activity in somatic cells, it was previously hypothesized that most cancers were characterized by telomere shortening in the early stage and on telomerase reactivation in the later stage of tumorigenesis (28). In some stem cell cancers, however, telomerase activity is already present, and reactivation is not required (5). In the current study, tumors developed from telomerase-positive B cells. Thus, it was surprising to find that TERT and telomerase activity were overexpressed in these tumors. In our study, there was no significant difference observed between the telomere lengths of tumors and normal tissues.
Recent studies have demonstrated telomere-independent functions of telomerase in cell cycle control and cell proliferation. TERT overexpression led to up-regulation of several proto-oncogenes including myb, growth factors, cell-fate regulators, and down-regulation of growth inhibitors (29). Conversely, down-regulation of TERT led to subsequent down-regulation of oncogenes and cell growth factors (30). Thus, it is possible that the myb overexpression observed in tumors with TERT integrations may be a consequence of TERT activation. TERT overexpression in epidermis increases embryonic stem cell (ESC) mobilization and proliferation in the absence of telomere-length changes (31). TERT also seems to enhance the survival of ESC by increasing the expression of stress response and defense genes (32). Recent studies show telomerase may also play a role in tumor progression and metastasis by activation of the glycolytic pathway and in suppression of tumor-cell differentiation (33, 34).
Telomerase reactivation is observed in >90% of human tumors but has not been seen in mouse tumors (35), suggesting that the role of telomerase in oncogenesis is different in mice than in humans (36). One possible reason for this difference is that the brief lifespan and telomere biology of rodent species is significantly different from humans. Mouse telomeres are long and do not shorten with aging (37). In contrast, avian telomeres shorten with age, and the pattern of telomerase expression in different tissues is more similar to humans (38). Chicken telomerase is active in early embryos and developing organs but is down-regulated in most somatic tissues unless they are transformed (39). The current study has shown that telomerase is frequently activated in chicken rapid-onset B cell lymphomas, as in human cancers, proving this to be an important and relevant model system for the study of telomerase involvement in oncogenesis.
Materials and Methods
Inverse PCR and Gene Identification.
The chicken tumors were generated in a study described in ref. 13. Tumor genomic DNA was prepared by standard proteinase K digestion followed by phenol-chloroform extraction. Inverse PCR was carried out as described in ref. 40 by using restriction enzyme Acc65I and L2/PolAcc65I-R primers (SI Table 2). Gene identity was analyzed with BLAST (National Center for Biotechnology Information) and BLAT (University of California, Santa Cruz, Genome Informatics) searches.
Luciferase Assay.
Secondary CEFs were transfected with various plasmid DNAs by using diethylaminoethyl (DEAE)-dextran as described in ref. 41. The pRL-CMV construct (Promega) was cotransfected into CEFs as a transfection control. Forty hours after transfection, CEFs were harvested and assayed for firefly and Renilla luciferase activities by using the Dual-Luciferase Reporter Assay System (Promega) while following the manufacturer's protocol. The luminescence was measured in the TopCount NXT microplate counter (Packard).
Southern Blot Analysis.
Genomic DNA was prepared with the DNeasy tissue kit (Qiagen) by following the manufacturer's protocol. Ten micrograms of genomic DNA were digested by EcoRI and separated on a 0.8% agarose gel. The TERT DNA probe was made by random labeling using a PCR fragment from TERT exon 2 as the template. The myb probe was made similarly from a plasmid containing a PCR fragment that was amplified with myb primers. The results were visualized by Typhoon 9410 Phosphorimager (Amersham Biosciences).
Real-Time RT-PCR.
Tumor RNA was extracted with RNA Bee (TEL-TEST) following the manufacturer's protocol and was treated with DNase I (Roche). Reverse transcription was carried out with random hexameric primers (Fermentas Life Sciences) and the iScript cDNA synthesis kit (Bio-Rad). Real-time PCR was performed using the iCycler iQ Multicolor Real-Time PCR Detection System (Bio-Rad), and the primers are listed in SI Table 1.
Telomerase Activity Assay.
The TRAP assay was carried out with the TRAPEZE Telomerase Detection Kit (Chemicon) following the manufacturer's protocol.
Telomere-Length Assay.
Ten micrograms of genomic DNA were digested with HaeIII and then were separated on a 0.8% agarose gel. The probe was made by end labeling of telomeric repeat DNA oligonucleotide 5′-(GGGATT)10G-3′ with polynucleotide kinase and gamma [32P]-ATP. The result was visualized by Typhoon 9410 Phosphorimager (Amersham Biosciences).
Supplementary Material
Acknowledgments
We thank Paul E. Neiman and the late Sandra J. Bowers (both at the Fred Hutchinson Cancer Research Center, Seattle, WA) for generation of the tumors; William Hayward (formerly at Memorial Sloan–Kettering Cancer Center, New York, NY) for the LR-9 and ΔLR-9 viral constructs and helpful discussions; Mohan Bolisetty for assistance; and the members of the K.L.B. Laboratory for their helpful comments. This work was supported by National Institutes of Health Grant R01 CA48746 from the National Cancer Institute (to K.L.B.). F.Y. is the recipient of the Millipore Foundation Dimitri V. d'Arbeloff Postdoctoral Fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709173104/DC1.
References
- 1.Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 2.Shay JW, Bacchetti S. Eur J Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 3.Neumann AA, Reddel RR. Nat Rev Cancer. 2002;2:879–884. doi: 10.1038/nrc929. [DOI] [PubMed] [Google Scholar]
- 4.Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, DePinho RA. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 5.Armanios M, Greider CW. Cold Spring Harb Symp Quant Biol. 2005;70:205–208. doi: 10.1101/sqb.2005.70.030. [DOI] [PubMed] [Google Scholar]
- 6.Kung HJ, Boerkoel C, Carter TH. Curr Top Microbiol Immunol. 1991;171:1–25. doi: 10.1007/978-3-642-76524-7_1. [DOI] [PubMed] [Google Scholar]
- 7.Akagi K, Suzuki T, Stephens RM, Jenkins NA, Copeland NG. Nucleic Acids Res. 2004;32:D523–D527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang W, Kanter MR, Dunkel I, Ramsay RG, Beemon KL, Hayward WS. J Virol. 1997;71:6526–6533. doi: 10.1128/jvi.71.9.6526-6533.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanter MR, Smith RE, Hayward WS. J Virol. 1988;62:1423–1432. doi: 10.1128/jvi.62.4.1423-1432.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clurman BE, Hayward WS. Mol Cell Biol. 1989;9:2657–2664. doi: 10.1128/mcb.9.6.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayward WS, Neel BG, Astrin SM. Nature. 1981;290:475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- 12.Payne GS, Bishop JM, Varmus HE. Nature. 1982;295:209–214. doi: 10.1038/295209a0. [DOI] [PubMed] [Google Scholar]
- 13.Polony TS, Bowers SJ, Neiman PE, Beemon KL. J Virol. 2003;77:9378–9387. doi: 10.1128/JVI.77.17.9378-9387.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pizer ES, Baba TW, Humphries EH. J Virol. 1992;66:512–523. doi: 10.1128/jvi.66.1.512-523.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neiman PE, Grbic JJ, Polony TS, Kimmel R, Bowers SJ, Delrow J, Beemon KL. Oncogene. 2003;22:1073–1086. doi: 10.1038/sj.onc.1206070. [DOI] [PubMed] [Google Scholar]
- 16.Greider CW, Blackburn EH. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 17.Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J, et al. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 18.Hosseini-Asl S, Modarressi MH, Atri M, Salhab M, Mokbel K, Mehdipour P. J Carcinog. 2006;5:17. doi: 10.1186/1477-3163-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim NW, Wu F. Nucleic Acids Res. 1997;25:2595–2597. doi: 10.1093/nar/25.13.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis RB, McClure J, Rup B, Niesel DW, Garry RF, Hoelzer JD, Nazerian K, Bose HR., Jr Cell. 1981;25:421–431. doi: 10.1016/0092-8674(81)90060-x. [DOI] [PubMed] [Google Scholar]
- 21.Hrdlickova R, Nehyba J, Liss AS, Bose HR., Jr J Virol. 2006;80:281–295. doi: 10.1128/JVI.80.1.281-295.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barr SD, Leipzig J, Shinn P, Ecker JR, Bushman FD. J Virol. 2005;79:12035–12044. doi: 10.1128/JVI.79.18.12035-12044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferber MJ, Montoya DP, Yu C, Aderca I, McGee A, Tnorland EC, Nagorney DM, Gostout BS, Burgart LJ, Boix L, et al. Oncogene. 2003;22:3813–3820. doi: 10.1038/sj.onc.1206528. [DOI] [PubMed] [Google Scholar]
- 24.Paterlini-Brechot P, Saigo K, Murakami Y, Chami M, Gozuacik D, Mugnier C, Lagorse D, Brechot C. Oncogene. 2003;22:3911–3916. doi: 10.1038/sj.onc.1206492. [DOI] [PubMed] [Google Scholar]
- 25.Trapp S, Parcells MS, Kamil JP, Schumacher D, Tischer BK, Kumar PM, Nair VK, Osterrieder N. J Exp Med. 2006;203:1307–1317. doi: 10.1084/jem.20052240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pizer E, Humphries EH. J Virol. 1989;63:1630–1640. doi: 10.1128/jvi.63.4.1630-1640.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weng NP, Granger L, Hodes RJ. Proc Natl Acad Sci USA. 1997;94:10827–10832. doi: 10.1073/pnas.94.20.10827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feldser DM, Hackett JA, Greider CW. Nat Rev Cancer. 2003;3:623–627. doi: 10.1038/nrc1142. [DOI] [PubMed] [Google Scholar]
- 29.Perrault SD, Hornsby PJ, Betts DH. Biochem Biophys Res Commun. 2005;335:925–936. doi: 10.1016/j.bbrc.2005.07.156. [DOI] [PubMed] [Google Scholar]
- 30.Kraemer K, Schmidt U, Fuessel S, Herr A, Wirth MP, Meye A. Int J Cancer. 2006;119:1276–1284. doi: 10.1002/ijc.21975. [DOI] [PubMed] [Google Scholar]
- 31.Flores I, Cayuela ML, Blasco MA. Science. 2005;309:1253–1256. doi: 10.1126/science.1115025. [DOI] [PubMed] [Google Scholar]
- 32.Armstrong L, Saretzki G, Peters H, Wappler I, Evans J, Hole N, von Zglinicki T, Lako M. Stem Cells. 2005;23:516–529. doi: 10.1634/stemcells.2004-0269. [DOI] [PubMed] [Google Scholar]
- 33.Bagheri S, Nosrati M, Li S, Fong S, Torabian S, Rangel J, Moore DH, Federman S, Laposa RR, Baehner FL, et al. Proc Natl Acad Sci USA. 2006;103:11306–11311. doi: 10.1073/pnas.0510085103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nosrati M, Li S, Bagheri S, Ginzinger D, Blackburn EH, Debs RJ, Kashani-Sabet M. Clin Cancer Res. 2004;10:4983–4990. doi: 10.1158/1078-0432.CCR-04-0134. [DOI] [PubMed] [Google Scholar]
- 35.Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 36.Akagi T. Trends Mol Med. 2004;10:542–548. doi: 10.1016/j.molmed.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 37.Carman TA, Afshari CA, Barrett JC. Exp Cell Res. 1998;244:33–42. doi: 10.1006/excr.1998.4207. [DOI] [PubMed] [Google Scholar]
- 38.Delany ME, Daniels LM, Swanberg SE, Taylor HA. Poult Sci. 2003;82:917–926. doi: 10.1093/ps/82.6.917. [DOI] [PubMed] [Google Scholar]
- 39.Delany ME, Daniels LM. Cytogenet Genome Res. 2003;102:309–317. doi: 10.1159/000075768. [DOI] [PubMed] [Google Scholar]
- 40.Tsuei DJ, Chen PJ, Lai MY, Chen DS, Yang CS, Chen JY, Hsu TY. J Virol Methods. 1994;49:269–284. doi: 10.1016/0166-0934(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 41.Arrigo S, Beemon K. Mol Cell Biol. 1988;8:4858–4867. doi: 10.1128/mcb.8.11.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.