Abstract
EmrE, a multidrug transporter from Escherichia coli, functions as a homodimer of a small four-transmembrane protein. The membrane insertion topology of the two monomers is controversial. Although the EmrE protein was reported to have a unique orientation in the membrane, models based on electron microscopy and now defunct x-ray structures, as well as recent biochemical studies, posit an antiparallel dimer. We have now reanalyzed our x-ray data on EmrE. The corrected structures in complex with a transport substrate are highly similar to the electron microscopy structure. The first three transmembrane helices from each monomer surround the substrate binding chamber, whereas the fourth helices participate only in dimer formation. Selenomethionine markers clearly indicate an antiparallel orientation for the monomers, supporting a “dual topology” model.
Keywords: membrane protein structure, multidrug transport, SMR family
One of the mechanisms by which cells neutralize the effect of toxic compounds is through the action of membrane transporters. Secondary transporters, such as the EmrE protein of Escherichia coli, couple the efflux of drugs to the inward movement of protons across the cell membrane (refs. 1 and 2, and references therein). EmrE is the prototypical member of the SMR (small multidrug resistance) family and is one of the smallest known transporters in nature, composed of only 110 amino acid residues. Studies have shown that the basic functional unit of EmrE is an oligomer, as would be expected for a membrane protein of its small size. It appears established that the basic functional unit of EmrE is a homodimer, as shown by oligomerization assays, substrate binding experiments, negative dominance studies, and crosslinking analyses (3–8). This conclusion is further supported by the existence of paired SMR proteins, such as YdgE/YdgF of E. coli, and EbrA/EbrB and YkkC/YkkD of Bacillus subtilis. These transporters require coexpression of the two component polypeptides for proper drug efflux activity and presumably form heterodimers analogous to the EmrE homodimer (9–13).
Electron microscopy (EM) studies of EmrE in complex with a transport substrate, tetraphenylphosphonium (TPP), and reconstituted in lipid bilayers have revealed the overall architecture of the transporter at 7.5 Å resolution in-plane and 16 Å perpendicular to the membrane (14). EmrE binds TPP as an asymmetric dimer, in a chamber that appears open to one side of the bilayer. Each monomer is composed of four transmembrane (TM) helices. Because of the low resolution, however, the monomers could not be delineated unambiguously, nor the TM segments assigned in a sequence-specific manner based on the experimental map alone.
We have previously reported x-ray crystal structures of EmrE in the unbound form and in complex with TPP (15, 16). Regrettably, the electron density maps were calculated in the wrong hand because of an unfortunate change of sign of the anomalous differences (17), and helices were misassigned in the TPP-bound model. Recalculation of the electron density map by using the proper sign of the anomalous differences and appropriate heavy atom site configuration resulted in maps with the correct hand. The inversion of the electron density map, however, does not change the topological relationships within the asymmetric unit; i.e., the relative orientation of the two monomers is a property that is unaffected by a mirror reflection through the unit cell origin. Thus, although certain findings of the original work regarding the chain trace were invalid, the conclusion of an antiparallel organization within the dimer is unchanged. We report here the corrected structures as recalculated from the original diffraction, new data from selenomethionine (SeMet)-labeled crystals, and functional assays of the recombinant proteins used for structure determination.
Results and Discussion
Expression, Purification, and Characterization of Recombinant EmrE.
N-terminally hexahistidine-tagged EmrE was produced either in vivo by overexpression in E. coli or in vitro using a cell-free translation system (see Materials and Methods). This tagged EmrE construct is functional when expressed in E. coli and conferred increased cellular resistance to positively charged cytotoxic compounds such as ethidium (2-fold above background), methyl viologen (>4-fold), and TPP (4-fold) (data not shown). Both in vivo- and in vitro-expressed EmrE were solubilized and purified in the detergent N-nonyl-β-d-glucoside (NG) and characterized for substrate binding activity by using fluorescence-based assays. Our experiments indicate that in vitro-expressed EmrE, which was an important source for SeMet-labeled protein, has essentially identical biochemical behavior to in vivo-expressed EmrE (see below).
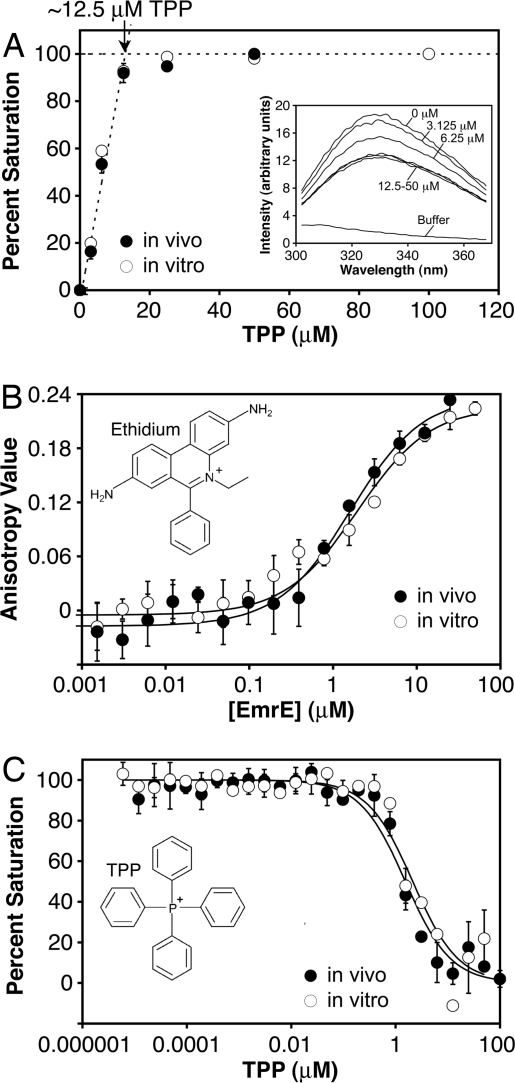
Binding of TPP to EmrE has been previously shown to quench fluorescence of the protein's tryptophan residues (18). As shown in Fig. 1A, the tryptophan emission intensity of EmrE displayed concentration-dependent quenching with added TPP (Inset). Saturation of 25 μM EmrE was observed at ≈12.5 μM TPP (intersection of the two dashed lines in Fig. 1A), in agreement with biochemical studies indicating a 2:1 protein:drug binding stoichiometry (6, 7). The data indicate no significant difference between the in vivo- and in vitro-derived proteins (note the near-superposition of the two curves in each of the three panels in Fig. 1).
Fig. 1.
Biochemical characterization of EmrE expressed in vivo (filled circles) and in vitro (open circles). The data shown are representative experiments performed in NG; error bars show the standard deviation of three replicates done in parallel. (A) Binding of EmrE to TPP, as assayed by tryptophan fluorescence quenching. (Inset) Typical emission spectra of 25 μM EmrE at the indicated concentrations of added TPP. Plotting the percent quenching at 320 nm vs. the added TPP shows saturation curves in the main panel. The total concentration of TPP binding sites was estimated from the intersection of the two dashed lines, indicated by the arrow (≈12.5 μM). (B) Binding of EmrE to ethidium, measured by fluorescence anisotropy. The data were fit to a simple binding model (see Materials and Methods). (C) Competition assays measuring TPP binding to EmrE. Note the near-superposition of the curves for in vivo and in vitro-expressed EmrE in all three panels.
Fluorescence anisotropy experiments, using ethidium as substrate, revealed dissociation constants (Kd) of 1.7 ± 0.4 and 1.8 ± 0.2 μM for in vivo and in vitro EmrE, respectively (Fig. 1B and Table 1). As expected, binding of EmrE to ethidium was competitively inhibited by TPP (Ki = 1.0 ± 0.2 μM for in vivo EmrE and 1.3 ± 0.2 μM for in vitro EmrE) (Fig. 1C and Table 1). Binding affinities were enhanced at least 5-fold for ethidium and 10-fold for TPP in the detergent N-dodecyl-β-d-maltoside (DDM) (Table 1), showing that the substrate binding activity of EmrE is influenced by the micellar environment, as noted previously (19). Despite the differences in binding affinity, however, the structure of the transporter is unchanged in these two detergents (discussed below and in Fig. 4). Overall, our measured affinities are in general agreement with published values (6, 7, 19–21) and show that the EmrE proteins used for these studies are functional in terms of substrate binding activity and stoichiometry.
Table 1.
Affinities of EmrE constructs for transport substrates
| Substrate | Detergent |
In vivo EmrE |
In vitro EmrE |
||||
|---|---|---|---|---|---|---|---|
| Affinity,† μM | SD‡ | n | Affinity,† μM | SD‡ | n | ||
| Ethidium | 0.3% NG | 1.69 | 0.43 | 13 | 1.79 | 0.18 | 9 |
| Ethidium | 0.1% DDM | 0.45 | 0.14 | 3 | 0.37 | 0.12 | 12 |
| TPP | 0.3% NG | 0.98 | 0.21 | 3 | 1.32 | 0.15 | 6 |
| TPP | 0.1% DDM | 0.08 | 0.02 | 6 | 0.05 | 0.02 | 9 |
†Affinity is expressed as Kd for ethidium and Ki for TPP, derived from fits of binding curves and competition curves, respectively.
‡Standard deviation of the indicated number of experiments.
Fig. 4.
Stereoview ribbon representation of the EmrE-TPP x-ray structure docked into the EM density map (14), contoured at 1.2σ. The TM helices are labeled, with the two monomers distinguished by asterisks. The density attributed to bound substrate in the EM map is indicated by the red arrow. This is in agreement with the As-derived position of TPP in the x-ray structure. The correspondence between the x-ray structure (derived from protein purified and crystallized in NG) and EM structure (purified in DDM and crystallized in reconstituted lipid bilayers) show that the tertiary and quaternary folds of the transporter are not distorted by the different detergent/lipid environments used.
Structure Determination of EmrE in the Absence of Ligand.
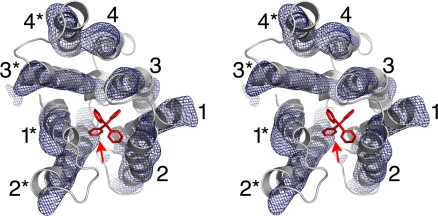
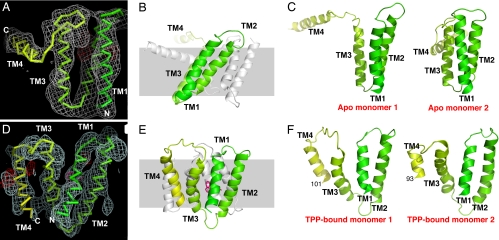
The unbound EmrE structure was recalculated from the original data by using anomalous diffraction from mercurial derivatives to a resolution limit of 4.5 Å [Fig. 2A, supporting information (SI) Fig. 5, and SI Table 2]. The asymmetric unit contained eight distorted EmrE monomers. As expected, each monomer is composed of four helices. The first three helices form a three-helix bundle, which packs against the equivalent helices of another EmrE molecule, forming a dimer (Fig. 2B). The fourth helices from each monomer interact with each other and project laterally from the main body of the dimer (Fig. 2 B and C), forming the bulk of the lattice contacts in this crystal (not shown). This EmrE conformation is difficult to reconcile with biochemical and other structural data. Given that apo EmrE was crystallized at pH 4, we believe that acid-induced partial denaturation of the protein may have resulted in a nonnative conformation that was stabilized by crystal contacts.
Fig. 2.
Structure determination of EmrE. (A) Experimental density for one apo EmrE monomer at 4.5-Å resolution. Anomalous Hg density (4σ), marking the positions of cysteine residues, is shown in red. The protein is shown in Cα trace and rendered in a color gradient, from green at the N terminus to yellow at the C terminus. (B) Ribbon representation of the distorted apo EmrE dimer. One monomer is rendered in color gradient with the helices labeled, and the other monomer is shown in gray. The approximate dimensions of a lipid bilayer are shown by the gray shading. (C) Views of the two apo EmrE monomers, with TM helices labeled. Note the extended configuration of the TM4 helices, which project away from the main body of the dimer. (D) Experimental density for one monomer of the EmrE-TPP complex at 3.8 Å (C2 crystal form), contoured at 1σ. Anomalous Se density (3σ) is shown in red. (E) Side view of the EmrE-TPP dimer (C2 form), with the dimensions of the lipid bilayer indicated. One monomer is colored in gradient and labeled, and the other is in gray. The bound TPP is colored red. Density for the colored monomer terminates at residue 105. (F) Views of the two monomers (P21 form), which are essentially the same as the C2 monomers, except for the shorter TM helices, which terminate at the indicated residues. Full-length EmrE has 110 amino acid residues. Note that the superhelical twists of TM1–3 are similar in the apo and TPP-bound forms but that the helix packing interactions and monomer–monomer interactions differ.
Structure Determination of EmrE Bound to TPP.
We determined the EmrE-TPP structure in three different crystal forms: a C2 form derived from in vivo-expressed EmrE and phased with anomalous data from the arsonium analogue of TPP to 4.0 Å resolution (original data), C2 crystals of in vitro EmrE phased with anomalous SeMet data at 3.8 Å, and a P21 form of in vitro SeMet-EmrE at 4.5 Å (SI Table 2). The three structures are essentially the same to the limit of the indicated resolutions but differ with regard to the extent of disorder at the termini. The C2 models include residues 6–105 for one monomer and 6–102 for the other (Fig. 2E). Because of greater disorder at the termini, the P21 model only includes residues 7–101 for one monomer and 7–93 for the other (Fig. 2F and SI Fig. 6).
Density for all helices in the dimer were clearly resolved, and helical connectivities were provided by visible loop densities (illustrated for one monomer in Fig. 2D). SeMet positions confirmed the identities and orientations of TM1 (residue 21) and TM4 (residues 91 and 92) for each monomer (Fig. 3). In addition, the SeMet sites, together with protruding densities for large aromatic regions (SI Fig. 7), provided the basis for assigning helical registry. Structure refinement statistics are shown in SI Table 2. The models were validated by weighted composite omit maps (SI Movies 1 and 2).
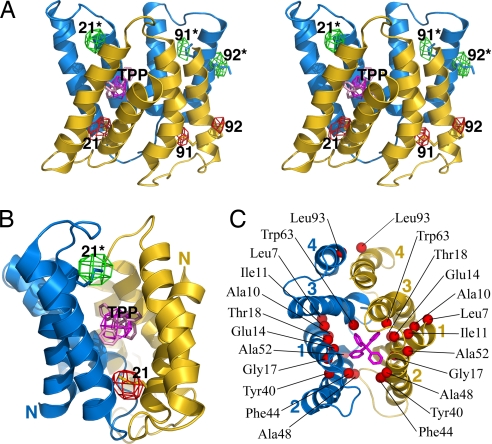
Fig. 3.
EmrE binds TPP as an antiparallel dimer. (A) Stereoview of the EmrE transporter in complex with TPP. The two monomers are colored blue and yellow, and the bound TPP is pink. Anomalous Fourier density from SeMet (colored red in one monomer and green in the other) and the arsonium analogue of TPP (magenta) are shown contoured at 3σ and 3.5σ, respectively. The TPP and SeMet residue positions are labeled, with the two monomers distinguished by asterisks. (B) “Front” view of the transporter, emphasizing the positions of SeMet markers in TM1. The N termini of the monomers are labeled. (C) “Top” view of the EmrE-TPP structure, with the TM helices labeled. Red spheres indicate the positions of residues that have been implicated in substrate binding and transport by biochemical and mutagenesis studies (18, 20, 23–28). The only residue removed from the binding chamber is Leu-93 (TM4). In the x-ray crystals, this residue appears to mediate lattice interactions across a twofold symmetry axis relating two dimers. This crystal packing interface was also observed in the two-dimensional crystals used to derive the EM structure of EmrE-TPP (14).
EmrE Binds TPP as an Antiparallel Homodimer.
The SeMet sites in the EmrE-TPP structure are related by a pseudo-twofold rotational axis running along the dimer interface parallel to the membrane plane. This dimer configuration is different from apo EmrE. As illustrated in Fig. 3A, the three Se sites in one monomer are positioned at the lower half of the transporter (protein is colored in yellow and Se density in red), whereas the Se sites in the other monomer are on the upper half (protein in blue, Se in green). The twofold relation between the monomers is more evident in Fig. 3B, which shows that the Se sites in the first TM helices (residues 21 and 21*) are located pseudo-symmetrically on opposite sides of the bilayer. These structural features clearly indicate that the x-ray structure of TPP-bound EmrE is an antiparallel dimer.
The two monomers adopt similar tertiary structures, with the two most highly conserved helices, TM1 and TM3, flanked by the less conserved TM2 and TM4 (Fig. 2F). Interestingly, the monomer fold does not contain an extensive hydrophobic core, suggesting that EmrE is an obligate dimer. The first three TM helices of the two monomers superimpose with an average root mean square deviation (rmsd) of 1.2 Å over equivalent helical Cα atoms (SI Fig. 8), whereas superposition of all four helices gives an average rmsd of 2.2 Å (not shown). The relative positions of TM1–3 are therefore more conserved than TM4. TM1–3 also have the same superhelical twist in apo and TPP-bound EmrE. Within the antiparallel dimer, the TM helices are arranged pairwise across the dimer interface (Fig. 3C). The bound substrate occupies a chamber surrounded by six helices, TM1, TM2, and TM3 from each monomer. Note that the substrate position is unambiguously defined in this structure, based on the anomalous signal from the arsonium analogue of TPP (the As density is colored magenta in Fig. 3 A and B). The two TM4 helices do not form part of the binding site and instead participate only in dimerization interactions, forming a four-helix bundle with the TM3 helices (Fig. 3C).
Comparison of the EmrE-TPP X-Ray Structure to the EM Structure.
The x-ray structure of EmrE-TPP closely matches the EM structure, which was determined from protein purified in the detergent DDM and crystallized in an artificial lipid bilayer (14). Rigid body fitting of EmrE-TPP into the EM density map reveals very good correspondence in the positions and tilts of the TM helices and location of the bound substrate (Fig. 4; also see SI Movie 3). Indeed, Tate and colleagues (14) have previously noted the presence of the same pseudo-twofold axis relating the two monomers and proposed an antiparallel dimer as one possible interpretation of the EM map. Note that the correspondence between the EM and x-ray structures indicates that the observed antiparallel arrangement of EmrE is not an artifact of crystallization in detergent, and that the structure of the transporter is not distorted by the different experimental methods used for purification and crystallization.
The TM assignment and topology of the x-ray structure matches a recent computational model based on the EM map, sequence conservation, and helical packing considerations based on the pseudo-twofold symmetry (22). Superposition of the x-ray helices with the computational model gives an average rmsd of 1.4 Å over equivalent Cα atoms (SI Fig. 9A). The rotational orientations of the TM helices are also similar, although the computational model varies somewhat with regards to the first TM helices (SI Fig. 9B). Overall, the x-ray structure confirms and validates the structure modeling approach used by Ben-Tal and colleagues (22).
Correlation of the EmrE-TPP Structure with Mutagenesis and Crosslinking Data.
Because of the limited resolution of the x-ray data, side chain conformations are not defined with atomic precision. Nevertheless, the Met residues are positioned with higher confidence based on the Se sites (Fig. 3 A and B), and some large aromatic side chains are resolved in the electron density maps (SI Fig. 7). With these markers, and because of the predominantly helical nature of EmrE, the locations of transmembrane residues may be positioned with some degree of certainty. With this limitation in mind, the x-ray structure of EmrE-TPP is in good agreement with studies that dissect the residue requirements for EmrE transport activity. For example, residues in the helical regions of TM1, TM2, and TM3 that have been shown to be important for TPP binding and transport activity generally map to the walls of the substrate binding chamber or interfaces between the surrounding helices (Fig. 3C) (18, 20, 23–28). Of particular note is Glu-14, which is absolutely conserved in homomeric SMR genes and is always present in at least one partner of paired SMR homologs (20, 24, 29). Based on the Se-derived position of Met-21, located two helical turns away, both Glu-14 residues point toward the binding chamber and appear well placed to form ionic contacts with the positively charged TPP molecule (Fig. 3C). This is consistent with biochemical and genetic studies showing that Glu-14 is absolutely required for substrate binding and proton-dependent transport (20, 24).
Also notable are mutations that either introduce or remove positively charged residues in loop regions of EmrE (29). These mutations are not expected to have any effect on the transmembrane packing or substrate transport mechanism per se. Nevertheless, they abolish transport activity, as shown by lack of growth resistance to ethidium upon expression of the EmrE mutants in E. coli (29). As discussed by the authors of the study, the mutations conferred unique Nin/Cin or Nout/Cout orientations on the EmrE constructs, whose lack of activity can be rescued or complemented by coexpression with the mutant construct of opposite topology (29).
The EmrE-TPP structure is not readily reconciled with a helical packing model, derived from thiol crosslinking data, showing that the two TM1 and two TM4 helices are in close contact within the dimer (5). This model is clearly at odds with the observed positions and orientations of these helices in the x-ray structure. Importantly, we note that TM1 and TM4 are the best-defined helices in the structure, based on the locations and grouping of the SeMet sites. The discrepancy may be partly explained by the observation, made first with LacY, that there is a clear tendency for crosslinks to underestimate distances, particularly in dynamic structures that fluctuate between different conformational states (30). It has also been previously suggested that a subset of the EmrE crosslinks could be explained by TM4-mediated tetramerization, about a twofold symmetry axis relating two dimers (22, 31). This twofold axis is observed in both the x-ray and EM structures (not shown).
A recent study indicates that EmrE dimers crosslinked at residue 108 are fully functional in substrate binding and transport activities (19). This biochemical finding has been interpreted as the definitive proof that EmrE could not possibly be an antiparallel dimer. This result may also be explained by TM4-mediated association between dimers, but a crosslinked tetramer was apparently ruled out by the authors of this study (19). In the x-ray structure of EmrE-TPP, the C-terminal ends of TM4, including residue 108, are disordered in both monomers. This is most pronounced in the P21 form, wherein the last 9 residues in one monomer and last 17 in the other are disordered (Fig. 2F). The extent and asymmetry of this disorder, combined with the observation that TM4 is not part of the TPP binding site, suggest that crosslinking of EmrE at position 108 (two residues from the terminus) may not necessarily disrupt the dimer structure or essential functional properties of the transporter in the detergent-solubilized state. Other speculative scenarios may be proposed, but we simply conclude that resolution between these conflicting models requires further empirical study.
The X-Ray Structure Supports a Dual Topology Model for EmrE.
The crystallographically observed antiparallel configuration of the EmrE dimer supports the conclusion that the EmrE protein is inserted in two orientations in the E. coli membrane. This contradicts a previous study by Schuldiner and coworkers (32), which suggested that EmrE has a unique Nin/Cin topology, but corroborates more recent studies from the von Heijne group (29, 33–35), which indicate that EmrE has dual topology.
Another crucial observation is that EmrE and other putative dual topology proteins belong to gene families, such as the SMR family, that include both singletons encoded by one gene and paired members encoded by a tandem of two genes (29, 35). Singleton SMR proteins like EmrE invariably have weak topology determinants, whereas paired SMR homologs harbor strong charge biases that favor opposite orientations in the membrane, as dictated by the “positive-inside rule.” The paired SMR proteins YdgE/YdgF of E. coli and EbrA/EbrB of B. subtilis have now been experimentally determined to have opposite topologies (i.e., YdgE and EbrA are Nout/Cout, whereas YdgF and EbrB are Nin/Cin) (12, 33, 34). These proteins, each of which show at least 65% sequence similarity and 30% identity to EmrE, have also been demonstrated to confer a multidrug resistant phenotype only when coexpressed with the corresponding partner (10, 11, 13). Similarly, the mutagenesis experiments by Rapp et al. (29) show that EmrE transport activity requires the coexpression of both Nin/Cin and Nout/Cout polypeptides, the two possible membrane orientations for EmrE (29). This “directed evolution” study demonstrated the conversion of EmrE from a homodimer to a “heterodimer” analogous to YdgEF and EbrAB. In a complementary study, Kikukawa et al. (36) reported the opposite result, converting EbrA and EbrB into homofunctional transporters. Specifically, when the topological determinants of either protein were altered or eliminated, theoretically allowing for construction of antiparallel homodimers, transport activity was observed in the absence of the native binding partner. As in the EmrE case, these manipulations left the substrate binding and transport determinants in the TM regions intact. We therefore submit that, on balance, the collective evidence favors an evolutionarily conserved architecture for the SMR family as antiparallel dimers, composed of either two copies of a dual topology protein or two distinct proteins of opposite topology.
Materials and Methods
Protein Expression and Purification.
EmrE was expressed either in vivo or in vitro with an N-terminal hexahistidine tag. The leader sequence is MGSSHHHHHHSSGLVPRGSH, where the italic residues comprise the thrombin cleavage site. Upon thrombin cleavage, the resulting construct is identical to the native form, except for three tag-derived residues (Gly-Ser-His) at the N terminus. EmrE was expressed in vivo by using E. coli BL21(DE3) transformed with a pET15b construct (EMD Biosciences). Cultures were induced overnight at 37°C with 2 mM IPTG added at mid-log phase (OD600 = 0.6). Expression levels were enhanced by oxygen depletion and addition of tetracycline to a final concentration of 10 μg/ml.
For purification, membrane proteins were extracted into 20 mM Tris (pH 8.0)/20 mM NaCl/2% (wt/vol) NG from either whole cells or membrane fractions derived from 100–200 g of cell paste. EmrE was then purified to homogeneity in a single step by using nickel-chelating chromatography in the same buffer but with 0.3% (wt/vol) NG. The polyhistidine tag was removed by proteolytic cleavage with thrombin, and the final construct was purified further by using ion-exchange and size-exclusion steps. The protein mass was verified to within 10 Da of expected (data not shown). EmrE proteins used for determination of the substrate-bound and apo forms were purified in 1 mM and 0 mM TPP, respectively.
SeMet-labeled EmrE was expressed by using a cell-free system and purified as described for the in vivo-expressed protein. The tagged EmrE coding sequence described above was cloned into pIVEX (Roche) for use as expression template. Lysates were produced from E. coli BL21(DE3) cells induced with IPTG (ref. 37 and references therein). Expression reactions contained the following components: 40% (vol/vol) lysate; 230 mM potassium glutamate; 58 mM Hepes (pH 7.5); 2% (wt/vol) PEG 8000; 80 mM ammonium acetate; 12 mM magnesium acetate; 1.2 mM ATP; 0.8 mM UTP, CTP, and GTP; 0.65 mM cAMP; 1.7 mM DTT; 34 μg/ml folinic acid; 30 mM 3-phosphoglycerate; 0.17 mg/ml E. coli tRNA; 4.5 mM SeMet; 2 mM each of the other 19 aa; and 50 μg/ml DNA template. The reaction mix was incubated at 30°C overnight with shaking. For large-scale production, the total volume of the reaction mix ranged from 20 to 100 ml. Upon purification, the measured mass of the labeled protein showed that four SeMet residues were incorporated per monomer, as expected from the sequence (data not shown).
Structure Determination.
The original diffraction intensities were converted to structure factors with the correct sign for the Friedel pairs. Data collection, phasing, and refinement statistics for each EmrE structure are shown in SI Table 2.
The apo EmrE structure was recalculated by using the original data derived from F222 crystals formed in 20 mM NaCl/20 mM sodium acetate (pH 4.0)/200–600 mM ammonium sulfate/15–30% (wt/vol) PEG 200/0.3–0.6% (wt/vol) NG. Phases were determined by using two-wavelength anomalous diffraction data collected from Hg-derivatized wild-type and C41S constructs. The program SNB (38) was used to determine the Hg substructure, and protein phases were combined by using PHASES (39). The asymmetric unit contained four sets of distorted dimers. Fourfold noncrystallographic symmetry (NCS) averaging, solvent flattening, and phase extension to 4.5-Å resolution were accomplished with PHASES (39) and CNS 1.2 (40). Electron density maps were traced manually by using CHAIN (41), and model refinement was performed by using CNS 1.2 with a maximum likelihood amplitude target function (mlf) (40). The topology of each monomer was confirmed by mercury-bound cysteine positions at residues 39, 41, and 95 (SI Fig. 5).
EmrE-TPP crystallized in 100–200 mM calcium chloride, 100 mM Tris (pH 6.4–7.2), 11–14% (wt/vol) PEG 2000 MME, and 0.3–0.6% (wt/vol) NG. The original data, indexed in C2, were derived from crystals of EmrE in complex with TPA, the arsonium analogue of TPP. The availability of new and better-quality crystals of SeMet-labeled protein from the cell-free system allowed us to obtain additional protein phases to facilitate the calculation of more accurate models and maps. SeMet-EmrE produced two distinct but related crystal forms of EmrE-TPP: a C2 form isomorphous with the EmrE-TPA crystals, and a P21 form. The data from the P21 crystals had a large pseudo-centering operator corresponding to the C centering in the C2 crystals. The C2 asymmetric unit contained one EmrE-TPP dimer, whereas the P21 form contained two (SI Fig. 6). As and Se sites were determined from two- or three-wavelength data sets by using SHELXD (42). Protein phases were independently determined for the C2-EmrE-TPA, C2-SeMet-EmrE-TPP, and P21-SeMet-EmrE-TPP crystal forms by using PHASES (39) and CNS 1.2 (40). Solvent flattening produced maps of sufficient quality for manual model building. Because of the controversy in the topology, each monomer was built independently with no NCS averaging within the dimer. A simulated annealing procedure (CNS 1.2) with a maximum likelihood phase probability distribution target function (mlhl) was used with hydrogen bonding restraints for helical residues (40). The models were validated by using SIGMAA-weighted composite omit maps with a mlf refinement target (CNS 1.2; see SI Movies 1 and 2).
Structural representations were rendered in PyMOL (DeLano Scientific). To properly reflect the low-resolution limit of the data, only Cα atoms were deposited in the Protein Data Bank.
Biochemical Characterization of EmrE Substrate Binding Activity.
Fluorescence anisotropy measurements were performed by using a Beckman DTX 880 fluorometer in the 384-well format, with excitation and emission filters set at 485 and 595 nm, respectively. Purified EmrE was serially diluted 2-fold into 30 μl of buffer [20 mM Tris (pH 8), 50 mM NaCl, 0.3% (wt/vol) NG or 0.1% (wt/vol) DDM] containing 1 μM ethidium. Measurements were repeated until samples reached equilibrium (typically <5 min). Anisotropy was calculated by using the equation A = (IV − G × IH)/(IV + 2G × IH), where A is the anisotropy value, IV is the fluorescence intensity polarized parallel to the excitation light, and IH is the fluorescence intensity polarized perpendicularly. The value of G was derived from the ratio of the parallel and perpendicular emission intensities of ethidium in the absence of EmrE (G = 0.59 in buffer only, 0.69 in buffer plus NG, and 0.72 in buffer plus DDM). This value represents the “intrinsic” anisotropy under the corresponding experimental conditions; the slightly higher values for buffers containing detergent reflect background partitioning of ethidium into micelles. Equilibrium binding isotherms were constructed by plotting the measured A values vs. the concentration of added EmrE. To determine Kd, the curve was fitted to the equation y = Ao + (Amax − Ao)([EmrE]/([EmrE] − Kd)), where Ao and Amax are the minimum and maximum anisotropy values, and [EmrE] is the concentration of added EmrE. Ao, Amax, and Kd were treated as free parameters.
Competition experiments used EmrE concentrations that achieved close to maximal binding of 1 μM ethidium (10 μM in NG buffer and 1 μM in DDM). TPP was serially diluted 2-fold into a 30-μl total volume and incubated for 30 min before anisotropy measurements. Curves were fitted to the equation y = Ao + (Amax − Ao)(1/(1 + 10log([TPP]/IC50)) to determine the IC50 value, treating Ao, Amax, and IC50 as free parameters. The inhibition constant was determined by using the equation Ki = IC50/(1 + [ethidium]/Kd), where Kd is the affinity for ethidium measured in parallel.
Tryptophan fluorescence measurements were performed by using a Varian Cary Eclipse scanning fluorometer in the 96-well format. Binding reactions contained 0.3% (wt/vol) NG or 0.1% (wt/vol) DDM, 25 μM EmrE, and TPP concentrations ranging from 0 to 100 μM in 100 μl of buffer. Each dilution was set-up in triplicate, and each replicate was scanned four times (excitation wavelength of 280 nm).
Supplementary Material
Acknowledgments
We thank T. Kudlicki and J. Fletcher (Invitrogen) for in vitro translation reagents; the staff at the Stanford Synchrotron Radiation Laboratory, Advanced Light Source, and Advanced Photon Source for assistance with data collection; and Z. Zhang, A. Ward, and J. Yu for technical help. The EM map was kindly provided by C. G. Tate (Medical Research Council, United Kingdom). This work was supported by National Institutes of Health Grants R01 GM67644 and Roadmap GM073197, National Aeronautics and Space Administration Grant NAG8-1834, the Skaggs Foundation for Chemical Biology, and Invitrogen. O.P. was supported by a postdoctoral fellowship from the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3B5D, 3B61, and 3B62).
This article contains supporting information online at www.pnas.org/cgi/content/full/0709387104/DC1.
References
- 1.Chung YJ, Saier MH., Jr Curr Opin Drug Discovery Dev. 2001;4:237–245. [PubMed] [Google Scholar]
- 2.Schuldiner S, Granot D, Mordoch SS, Ninio S, Roten D, Soskine M, Tate CG, Yerushalmi H. News Physiol Sci. 2001;16:130–134. doi: 10.1152/physiologyonline.2001.16.3.130. [DOI] [PubMed] [Google Scholar]
- 3.Yerushalmi H, Lebendiker M, Schuldiner S. J Biol Chem. 1996;271:31044–31048. doi: 10.1074/jbc.271.49.31044. [DOI] [PubMed] [Google Scholar]
- 4.Rotem D, Sal-man N, Schuldiner S. J Biol Chem. 2001;276:48243–48249. doi: 10.1074/jbc.M108229200. [DOI] [PubMed] [Google Scholar]
- 5.Soskine M, Steiner-Mordoch S, Schuldiner S. Proc Natl Acad Sci USA. 2002;99:12043–12048. doi: 10.1073/pnas.192392899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tate CG, Ubarretxena-Belandia I, Baldwin JM. J Mol Biol. 2003;332:229–242. doi: 10.1016/s0022-2836(03)00895-7. [DOI] [PubMed] [Google Scholar]
- 7.Butler PJ, Ubarretxena-Belandia I, Warne T, Tate CG. J Mol Biol. 2004;340:797–808. doi: 10.1016/j.jmb.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Elbaz Y, Steiner-Mordoch S, Danieli T, Schuldiner S. Proc Natl Acad Sci USA. 2004;101:1519–1524. doi: 10.1073/pnas.0306533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jack DL, Storms ML, Tchieu JH, Paulsen IT, Saier MH., Jr J Bacteriol. 2000;182:2311–2313. doi: 10.1128/jb.182.8.2311-2313.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masaoka Y, Ueno Y, Morita Y, Kuroda T, Mizushima T, Tsuchiya T. J Bacteriol. 2000;182:2307–2310. doi: 10.1128/jb.182.8.2307-2310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishino K, Yamaguchi A. J Bacteriol. 2001;183:5803–5812. doi: 10.1128/JB.183.20.5803-5812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kikukawa T, Miyauchi S, Araiso T, Kamo N, Nara T. Biochem Biophys Res Commun. 2007;358:1071–1075. doi: 10.1016/j.bbrc.2007.05.032. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, Ma C, Pornillos O, Xiu X, Chang G, Saier MH., Jr Biochemistry. 2007;46:5218–5225. doi: 10.1021/bi7001604. [DOI] [PubMed] [Google Scholar]
- 14.Ubarretxena-Belandia I, Baldwin JM, Schuldiner S, Tate CG. EMBO J. 2003;22:6175–6181. doi: 10.1093/emboj/cdg611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma C, Chang G. Proc Natl Acad Sci USA. 2004;101:2852–2857. doi: 10.1073/pnas.0400137101. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Pornillos O, Chen YJ, Chen AP, Chang G. Science. 2005;10:1950–1953. doi: 10.1126/science.1119776. [DOI] [PubMed] [Google Scholar]
- 17.Chang G, Roth CB, Reyes CL, Pornillos O, Chen YJ, Chen AP. Science. 2006;314:1875. doi: 10.1126/science.314.5807.1875b. [DOI] [PubMed] [Google Scholar]
- 18.Elbaz Y, Tayer N, Steinfels E, Steiner-Mordoch S, Schuldiner S. Biochemistry. 2005;44:7369–7377. doi: 10.1021/bi050356t. [DOI] [PubMed] [Google Scholar]
- 19.Soskine M, Mark S, Tayer N, Mizrachi R, Schuldiner S. J Biol Chem. 2006;281:36205–36212. doi: 10.1074/jbc.M607186200. [DOI] [PubMed] [Google Scholar]
- 20.Muth TR, Schuldiner S. EMBO J. 2000;19:234–240. doi: 10.1093/emboj/19.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sikora CW, Turner RJ. Biophys J. 2005;88:475–482. doi: 10.1529/biophysj.104.049247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleishman SJ, Harrington SE, Enosh A, Halperin D, Tate CG, Ben-Tal N. J Mol Biol. 2006;364:54–67. doi: 10.1016/j.jmb.2006.08.072. [DOI] [PubMed] [Google Scholar]
- 23.Mordoch SS, Granot D, Lebendiker M, Schuldiner S. J Biol Chem. 1999;274:19480–19486. doi: 10.1074/jbc.274.27.19480. [DOI] [PubMed] [Google Scholar]
- 24.Yerushalmi H, Schuldiner S. J Biol Chem. 2000;275:5264–5269. doi: 10.1074/jbc.275.8.5264. [DOI] [PubMed] [Google Scholar]
- 25.Yerushalmi H, Mordoch SS, Schuldiner S. J Biol Chem. 2000;276:12744–12748. doi: 10.1074/jbc.M010979200. [DOI] [PubMed] [Google Scholar]
- 26.Gutman N, Steiner-Mordoch S, Schuldiner S. J Biol Chem. 2003;278:16082–16087. doi: 10.1074/jbc.M213120200. [DOI] [PubMed] [Google Scholar]
- 27.Sharoni M, Steiner-Mordoch S, Schuldiner S. J Biol Chem. 2005;280:32849–32855. doi: 10.1074/jbc.M504910200. [DOI] [PubMed] [Google Scholar]
- 28.Rotem D, Steiner-Mordoch S, Schuldiner S. J Biol Chem. 2006;281:18715–18722. doi: 10.1074/jbc.M602088200. [DOI] [PubMed] [Google Scholar]
- 29.Rapp M, Seppälä S, Granseth E, von Heijne G. Science. 2007;315:1282–1284. doi: 10.1126/science.1135406. [DOI] [PubMed] [Google Scholar]
- 30.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 31.Rath A, Melnyk RA, Deber CM. J Biol Chem. 2006;281:15546–15553. doi: 10.1074/jbc.M600434200. [DOI] [PubMed] [Google Scholar]
- 32.Ninio S, Elbaz Y, Schuldiner S. FEBS Lett. 2004;562:193–196. doi: 10.1016/S0014-5793(04)00240-6. [DOI] [PubMed] [Google Scholar]
- 33.Daley DO, Rapp M, Granseth E, Melen K, Drew D, von Heijne G. Science. 2005;308:1321–1323. doi: 10.1126/science.1109730. [DOI] [PubMed] [Google Scholar]
- 34.Granseth E, Daley DO, Rapp M, Melen K, von Heijne G. J Mol Biol. 2005;352:489–494. doi: 10.1016/j.jmb.2005.07.053. [DOI] [PubMed] [Google Scholar]
- 35.Rapp M, Granseth E, Seppälä S, von Heijne G. Nat Struct Mol Biol. 2006;13:112–116. doi: 10.1038/nsmb1057. [DOI] [PubMed] [Google Scholar]
- 36.Kikukawa T, Nara T, Araiso T, Miyauchi S, Kamo N. Biochim Biophys Acta. 2006;1758:673–679. doi: 10.1016/j.bbamem.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 37.Sitaraman K, Esposito D, Klarmann G, Le Grice SF, Hartley JL, Chatterjee DK. J Biotechnol. 2004;110:257–263. doi: 10.1016/j.jbiotec.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 38.Miller R, Gallo SM, Khalak HG, Weeks CM. J Appl Crystallogr. 1994;27:613–621. [Google Scholar]
- 39.Furey W, Swaminathan S. Methods Enzymol. 1997;277:590–620. doi: 10.1016/s0076-6879(97)77033-2. [DOI] [PubMed] [Google Scholar]
- 40.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 41.Sack JS. J Mol Graphics. 1988;6:224–225. [Google Scholar]
- 42.Schneider TR, Sheldrick GM. Acta Crystallogr D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.