Abstract
How are the rates of aging of different tissues coordinated? In Caenorhabditis elegans, decreasing insulin/IGF-1 signaling extends lifespan by activating the transcription factor DAF-16/FOXO. If DAF-16 levels are experimentally increased in one tissue, such as the intestine, DAF-16 activity in other tissues rises. Here we test the hypothesis that this “FOXO-to-FOXO” signaling occurs via feedback regulation of ins-7 insulin gene expression. We find that DAF-16 regulates ins-7 expression in the intestine, and that preventing this regulation blocks FOXO-to-FOXO signaling from the intestine to other tissues. Our findings show that feedback regulation of insulin gene expression coordinates DAF-16 activity among the tissues, and they establish the intestine, which is the animal's entire endoderm, as an important insulin-signaling center.
Keywords: aging, DAF-2, DAF-16, FOXO
In Caenorhabditis elegans, reducing the activity of the insulin/IGF-1-receptor DAF-2 doubles lifespan (1). A striking feature of these long-lived animals is that, throughout their lives, they closely resemble younger wild-type animals (2, 3). In other words, relative to one another, the rates of aging of the different tissues appear normal. In principle, this apparent normalcy could arise from a similar cell-autonomous reduction of insulin/IGF-1 action in all tissues. However, genetic mosaic analysis indicates that signaling between the different tissues of the animal, downstream of insulin/IGF-1 receptor activity, plays an important role (4, 31). For example, removing the daf-(+) gene from an early blastomere that produces only some of the tissues extends the lifespan of the entire animal, keeping wild-type tissues alive long after they would normally die (4). Thus cells that lack daf-2 receptor activity not only change their own rates of aging, they also signal wild-type cells to change their rates of aging as well.
How might daf-2 activity influence downstream intercellular signaling? The insulin/IGF-1 pathway affects aging, at least in part, by controlling the activity of a phosphoinositol 3-kinase/PDK/AKT-kinase cascade that phosphorylates the FOXO-family transcription factor DAF-16 and prevents DAF-16 accumulation in the nucleus (5–12). daf-16 is required for the longevity of daf-2 mutants. When insulin/IGF-1 signaling levels fall, DAF-16 accumulates in nuclei, where it changes the expression of downstream longevity genes (13, 32–34), including, presumably, genes encoding downstream intercellular signals. Consistent with this idea, expression of daf-16 only in the intestine or neurons of daf-2 mutants is sufficient to increase the lifespan of the entire animal (14).
The insulin/IGF-1 pathway influences lifespan cell nonautonomously in flies and mice as well as worms. For example, increasing dFOXO activity only in fat tissue extends the lifespan of flies (15, 16), and loss of the insulin receptor in the fat tissue, which would be predicted to increase FOXO activity, extends the lifespan of mice (17). The intestine of C. elegans functions as the animal's entire endoderm, including its site of fat storage. Thus identifying the downstream longevity signals regulated by insulin/IGF-1 signaling in the intestine of C. elegans could have general significance.
Genetic experiments indicate that signals regulated by DAF-16 in one tissue affect the activity of DAF-16 in responding cells (14). If daf-16 expression is increased in a single tissue, such as the intestine (using a tissue-specific promoter), expression of the direct DAF-16-target gene sod-3 (superoxide dismutase) (18, 19) is increased not only in that tissue but in other tissues as well. This increased sod-3 expression requires DAF-16 in responding cells, because it is not observed in a daf-16(−) background (14). We call this communication “FOXO-to-FOXO” signaling and hypothesize that it helps to equalize the activity of DAF-16 in the different tissues of the animal, thereby helping to coordinate their rates of aging.
How might DAF-16 activity in one tissue influence DAF-16 activity elsewhere? Because DAF-16 is known to be regulated by insulin/IGF-1 signaling, an attractive model is that FOXO-to-FOXO signaling is mediated by insulin-like genes whose expression is regulated by DAF-16 (4). In this scenario, changing DAF-16 activity in one tissue would change insulin gene expression in that tissue, which in turn would alter extracellular insulin levels and ultimately the activity of DAF-16/FOXO in other tissues. Recently, we identified a number of genes encoding putative signaling molecules in microarrays of daf-2 and daf-16 mutants (13). Among these was an insulin-like gene, ins-7. In this study, we tested the hypothesis that INS-7 is a signal that is regulated by DAF-16 to influence FOXO-to-FOXO signaling. Together our findings indicate that this is the case, and in addition they establish the endoderm of C. elegans as an important insulin-signaling center in the animal.
Results
ins-7 Activates the DAF-2 Pathway.
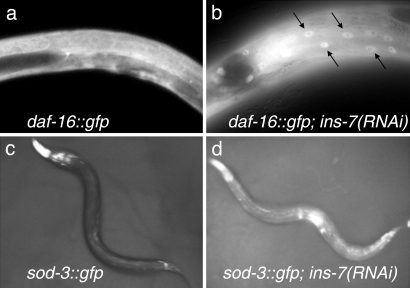
The first step in testing the hypothesis that ins-7 influences FOXO-to-FOXO signaling was to determine the normal function of ins-7 in the animal. ins-7 RNAi or mutation extends the lifespan of wild-type adults and increases the penetrance of a daf-2 loss-of-function developmental phenotype, the formation of dauer larvae, suggesting that INS-7 is a DAF-2 agonist [ref. 13 and supporting information (SI) Fig. 5]. If this is the case, then ins-7 RNAi should increase the nuclear accumulation of DAF-16 and the expression of the DAF-16 target gene sod-3. We found that ins-7 RNAi stimulated nuclear localization of a DAF-16::GFP protein fusion (Fig. 1 a and b) (6). In addition, ins-7 RNAi increased Psod-3::gfp expression (Fig. 1 c and d). Thus ins-7 RNAi likely promotes longevity and dauer formation by inhibiting DAF-2 and activating DAF-16.
Fig. 1.
ins-7 influences targets of the DAF-2 insulin/IGF-1 pathway. (a) Wild-type animals expressing Pdaf-16::daf-16::gfp cultured on control RNAi bacteria exhibit diffuse DAF-16::GFP localization. (b) Subjecting these animals to ins-7 RNAi increases DAF-16::GFP nuclear localization (arrows). (c) Wild-type animals expressing Psod-3::gfp and grown on control RNAi bacteria exhibit low levels of GFP in the body and higher levels in the head region (light areas). (d) ins-7 RNAi increases Psod-3::gfp expression throughout the body.
DAF-2 and DAF-16 Regulate ins-7 Expression in the Intestine.
Many of the ≈38 insulin-like genes in C. elegans are expressed in the nervous system (20). In addition, killing specific sensory neurons or inhibiting their functions genetically triggers DAF-16 nuclear localization (9) and extends lifespan (21). Moreover, signals that regulate dauer formation change the expression of certain insulin-like genes in sensory neurons (22). For this reason, it is widely believed that the biologically relevant site of insulin production is primarily the nervous system, despite the fact that expression of insulin-like genes has been observed in other tissue types (20).
In genetic experiments, the intestine behaves as a potent source of FOXO-to-FOXO signals; in fact, DAF-16 activity in the intestine appears more effective at elevating DAF-16 activity in other tissues than is DAF-16 activity in neurons (14). Therefore, it was of great interest to learn whether ins-7 was regulated by DAF-2 and DAF-16 in the intestine.
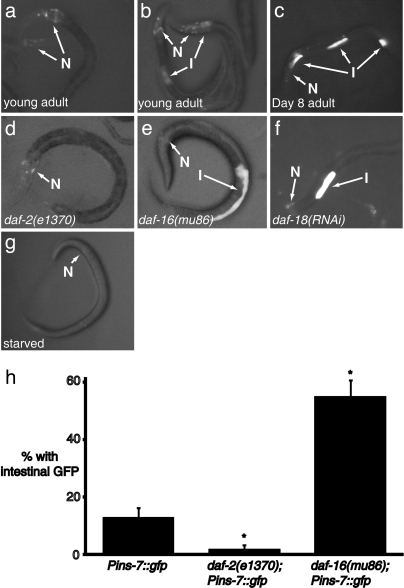
Previously, Pins-7::gfp was reported to be expressed exclusively in neurons (20). However, we observed that some animals carrying Pins-7::gfp (≈12%) also expressed Pins-7::gfp in intestinal cells (Fig. 2 b and h). In addition, intestinal (but not neuronal) Pins-7::gfp expression increased with age (Fig. 2c). On day 8 of adulthood, ≈30% of animals exhibited intestinal Pins-7::gfp expression. We note that not every intestinal cell expressed Pins-7::gfp in these animals. Instead, apparently random subsets of intestinal cells expressed the transgene in different animals. This expression pattern may reflect the endogenous expression pattern of ins-7; however, similar random subintestinal expression patterns have been seen with other intestinal promoter fusions, such as fusions to the ges-1 (gut-esterase) promoter, suggesting that the random pattern may be a more general property of transgenes expressed in the intestine.
Fig. 2.
Regulation of Pins-7::gfp expression. I, intestinal cell; N, neurons. (a–g) Pins-7::gfp expression under different conditions. (a) Typical young adult, exhibiting neuronal but not intestinal gfp expression; (b) young adult expressing Pins-7::gfp in intestinal cells; (c) typical day 8 adult, exhibiting intestinal as well as neuronal gfp expression (d) typical Pins-7::gfp; daf-2(e1370) mutant, with no intestinal expression (see h); (e) typical Pins-7::gfp; daf-16(mu86) mutant, with pronounced intestinal expression; (f) typical Pins-7::gfp animal subjected to daf-18/PTEN RNAi; and (g) typical starved wild-type animal, with sharply reduced ins-7::gfp expression; (h) percentage of wild-type (n = 100) and mutant Pins-7::gfp worms (n = 84 for daf-16, n = 102 for daf-2) expressing GFP fluorescence in intestinal cells. Standard error of the proportion (SEP). A Z test for two proportions shows that daf-2 and daf-16 mutants are each significantly different from the vector control at the 99% confidence level (*).
If intestinal ins-7 mediates FOXO-to-FOXO signaling, then intestinal ins-7 levels should respond to changes in daf-2 or daf-16 activity. We reduced daf-16 activity with the daf-16 null mutation mu86 and observed a striking increase in intestinal Pins-7::gfp expression in young adults (to 55%; Fig. 2 e and h; P < 0.01). In contrast, there was no obvious effect of daf-16(mu86) on neuronal Pins-7::gfp expression (Fig. 2e). We observed a similar increase in intestinal Pins-7::gfp expression in daf-18/PTEN mutants, which have increased insulin/IGF-1 signaling levels because they lack a phosphatase that counteracts PI 3-kinase activity (Fig. 2f).
We reduced daf-2 activity with daf-2(e1370) and observed a reduction Pins-7::gfp in the intestine to 2% (P < 0.01) but not in the nervous system (Fig. 2 d and h). Together, these findings suggested that the intestine is the primary tissue in which ins-7 expression is regulated by insulin/IGF-1 signaling.
ins-7 Likely Mediates FOXO-to-FOXO Signaling.
Because ins-7 was regulated by DAF-2 and DAF-16 in the intestine, it was a candidate for a signal that influences FOXO-to-FOXO signaling from the intestine to other tissues. To test directly whether this was the case, we increased DAF-16 levels in the intestine but prevented this increase from decreasing ins-7 expression.
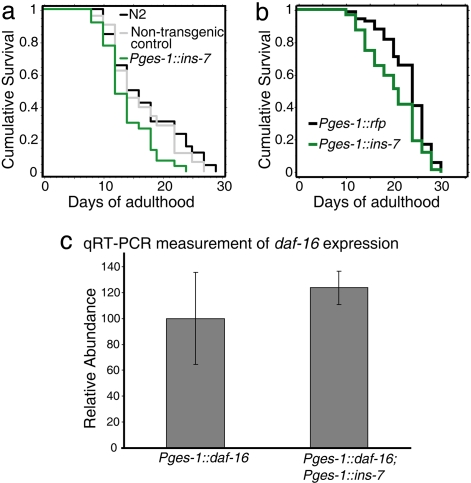
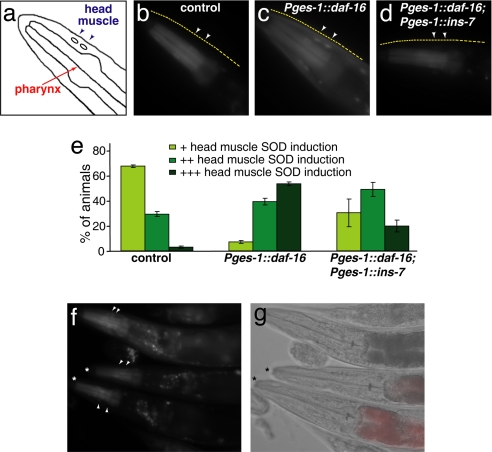
To insulate ins-7 gene expression from the influence of DAF-16, we fused the ins-7 coding region to the promoter of the intestinal gene ges-1. Animals expressing this transgene had a slightly shorter lifespan than did wild-type animals or control animals expressing Pges-1::rfp (Fig. 3 a and b), as predicted if the transgene increases INS-7 levels in the animal. We then increased the level of DAF-16 in the intestine using a functional Pges-1::daf-16::gfp fusion and assayed the level of sod-3::gfp in hypodermis and muscle. In control animals lacking the ins-7 transgene, elevating intestinal DAF-16 levels increased Psod-3::gfp expression levels in other tissues. (Compare Fig. 4b, in which only the pharynx expresses gfp, with Fig. 4c, in which many additional tissues express gfp.) However, when intestinal DAF-16 levels were increased in animals carrying Pges-1:: ins-7, little or no increase in DAF-16::GFP was observed in other tissues (Fig. 4 d and e). (Note the similarity of the animal in Fig. 4d to the wild-type control in Fig. 4b.)
Fig. 3.
Intestinally expressed ins-7 shortens lifespan. (a) Mean lifespan of wild-type (N2) = 17.0 ± 1.2, non-transgenic control = 16.3 ± 1.3, and Pges-1:ins-7 = 13.9 ± 0.7, P = 0.0096 (non-transgenic vs. Pges-1::ins-7). (b) Mean lifespan of Pges-1::gfp = 23.0 ± 0.6, Pges-1:ins-7 = 20.1 ± 0.6, P = 0.0007. See SI Table 1 for additional trials and statistical analysis. (c) Intestinally expressed ins-7 does not affect daf-16 expression. Quantitative RT-PCR of intestinally expressed daf-16 in wild-type and Pges-1::ins-7 backgrounds. Results are from two biological repeats; error bars represent standard deviation.
Fig. 4.
Inhibiting DAF-16 regulation of ins-7 decreases the ability of intestinal DAF-16 to affect DAF-16 activity in other tissues. All animals carry Psod-3::gfp. (a) Schematic illustration of the animal's head. (b–d) The upper edge of the animal's head is indicated by the dashed yellow line, and arrowheads point to muscle nuclei. (b) Control wild-type animals express Psod-3::gfp in the pharynx but not in surrounding head tissue. (c) Increasing intestinal DAF-16 levels by using Pges-1::daf-16 increases Psod-3::gfp expression throughout the head. [This increased Psod-3::gfp expression requires increased DAF-16 activity in nonintestinal tissues, because it is not seen in a daf-16(−) background (14).] (d) Constitutive expression of ins-7 from the ges-1 intestinal promoter (Pges-1::ins-7) reduces the ability of intestinal Pges-1::daf-16 to stimulate Psod-3::gfp expression in the head. (e) Semiquantification for b–d (n ≥ 109). (f and g) Pges-1::daf-16 animals with (*) and without Pges-1::ins-7. Animals carrying Pges-1::ins-7 have red intestinal fluorescence from the coinjected Pges-1::rfp construct (see Materials and Methods and SI Fig. 6).
We considered the possibility that Pges-1::ins-7 might reduce expression of intestinal Pges-1::daf-16 by titrating factors required for Pges-1 promoter activity, thereby decreasing FOXO-to-FOXO signaling for trivial reasons. However, quantitative RT-PCR experiments indicated this was not the case (Fig. 3c). Thus these findings suggest that DAF-16 activity in the intestine activates DAF-16 elsewhere in the animal, at least in part, by down-regulating intestinal expression of ins-7.
Discussion
In principle, a tissue's rate of aging might be specified in a purely cell-autonomous fashion, for example, by the rate of production of reactive oxygen species generated within its own mitochondria. However, the finding that expressing components of the insulin/IGF-1 longevity pathway in subsets of cells can affect the rate of aging of the entire organism implies the existence of a mechanism that actively coordinates the rates of aging between the different tissues. In this study, we have shown that in C. elegans, this communication between the tissues is mediated, at least in part, by insulin-like peptides. Specifically, we showed that the ins-7 gene is regulated by DAF-16 activity in the intestine, and that this regulation, in turn, allows DAF-16 activity in the intestine to influence DAF-16 activity in other tissues.
Feedback Regulation of ins-7 Expression.
Our attention was first drawn to ins-7 as a possible mediator of FOXO-to-FOXO signaling because its expression changed in DAF-2-pathway microarrays (13). ins-7 is expressed in two tissues, the nervous system and the intestine; however, insulin/IGF-1 signaling appears to regulate expression of ins-7 primarily in the intestine. Because ins-7 functions as expected for a DAF-2 agonist, and because its expression is activated by the insulin/IGF-1-response pathway in the intestine, ins-7 is a part of a positive-feedback (i.e., feed-forward) regulatory loop that would be predicted to amplify upward or downward fluctuations in insulin/IGF-1 signaling.
In C. elegans, the intestine appears to behave as the animal's entire endoderm. For example, the intestine is the animal's site of fat storage (adipose tissue) and of yolk production (liver). These studies suggest the C. elegans intestine may also perform functions of the mammalian pancreas, also an endodermal organ. In fact, the insulin response pathway is known to up-regulate insulin gene expression in the mammalian pancreas (23), just as the insulin-response pathway up-regulates ins-7 in the C. elegans intestine. Consistent with this, ins-7 expression dropped sharply in the intestine (as well as the nervous system) under fasting conditions (Figs. 2g).
ins-7 Behaves as Expected for a Signal That Influences FOXO-to-FOXO Signaling.
The main goal of this study was to test the hypothesis that DAF-16 activity in the intestine influences DAF-16 activity in other tissues by feedback regulation of insulin gene expression. The finding that DAF-16 regulated the expression of ins-7 within the intestine satisfied one prediction of this hypothesis. However, it was important to test the significance of this feedback regulation directly, because the effects of changes in the expression of a small subset of insulin genes might be masked by the constitutive expression of many other insulin-like genes in the animal.
We tested our hypothesis by preventing DAF-16 from down-regulating ins-7 gene expression. We found that constitutive intestinal ins-7 expression prevented elevated levels of intestinal DAF-16 from triggering muscle and hypodermal expression of Psod-3::gfp. The simplest interpretation of this finding is that our model was correct: down-regulation of ins-7 in the intestine by DAF-16 lowers INS-7 levels in the animal, which in turn triggers DAF-16 activity in muscle and hypodermis. Because we know that DAF-16 does down-regulate ins-7 expression in the intestine, and that lowering INS-7 levels in the animal reduces DAF-2 pathway activity, this interpretation is clearly the most straightforward. However, we cannot rule out the possibility that, as with any overexpression experiment, artificially high levels of a protein, in this case INS-7, bypasses the activity of the normal signaling system. Thus, we conclude that ins-7 can function in FOXO-to-FOXO signaling and most likely does so in normal animals (Fig. 4).
INS-7 may not be the only insulin-like peptide involved in FOXO-to-FOXO signaling. In our microarray analysis and subsequent experiments (ref. 13; unpublished data), we found that the gene ins-18 is also feedback-regulated by DAF-16 in the intestine, but in the opposite direction from ins-7. An RNAi clone predicted to target ins-18 prevented DAF-16 nuclear localization, shortened lifespan and prevented dauer formation, suggesting that it targeted a DAF-2 antagonist. This RNAi clone also completely prevented FOXO-to-FOXO signaling from the intestine to other tissues (SI Fig. 7). It is possible that this clone targeted more than one insulin-like gene, because most of these phenotypes were not observed with an ins-18 deletion mutant (data not shown). These findings raise the possibility that insulin-like genes that behave as DAF-2 antagonists rather than agonists may also contribute to FOXO-to-FOXO signaling.
Positive-Feedback Loops May Contribute to Cellular Consensus Mechanisms.
Feed-forward regulatory loops should, in principle, continue to amplify small perturbations until the system has reached a state of maximal or minimal activity. This may occur during dauer formation. When exposed to mild dauer-inducing conditions, some worms in a population become dauers, whereas others become adults. Within each animal, all cells reach a consensus and adopt the same (dauer vs. adult) fate. A positive-feedback system involving intercellular signals such as the one described here is likely to facilitate this decision process. In the regulation of lifespan, there must be factors that limit the ability of this system to spiral endlessly up or down, because many insulin-pathway mutations have intermediate effects on lifespan.
Insulin-like peptides cannot be the only signals that act downstream of DAF-16 to influence lifespan. When daf-16 is expressed only within the intestine of daf-16; daf-2 double mutants, lifespan is increased ≈60% (14). In these animals, tissues that would normally die much earlier, such as muscles and neurons, remain alive, because the long-lived animals continue to move. Because these nonintestinal tissues do not contain DAF-16, another signaling pathway downstream of DAF-16 must also influence aging. The secreted peptide scl-1 is a candidate for such a downstream signal (24), as are other predicted signaling molecules whose gene expression changes in DAF-16 in microarrays (13, 32).
In summary, we have shown that an important mechanism by which the activity of the DAF-2/DAF-16 signaling pathway is coordinated among the different tissues of the animal is through positive-feedback regulation of an insulin-like peptide. This feedback regulation has the effect of equalizing DAF-16 activity in the different tissues, which would be expected to bring the rates of aging of the different tissues into alignment. In worms as well as flies, the nervous system has been regarded as the primary site of insulin production, despite the fact that insulin-like genes can be expressed in non-neuronal tissues in both organisms (20, 35). This study shows that, at least in C. elegans, the intestine/endoderm is an important site of insulin production and regulation.
In Drosophila, overexpressing dFOXO in the pericerebral fat body decreases expression of the insulin gene dilp2 in insulin-producing neurons (15). In this case, feedback regulation of insulin-like gene expression is taking place, but across two different cell types. The signaling pathway by which dFOXO activity in fat tissue communicates with neurons is unknown. It will be interesting to learn whether, as in worms, this signaling is mediated by insulin-like peptides produced by fat tissue. Interestingly, dFOXO may play a key role on the receiving end of this pathway, because dFOXO has been shown to act within neurons to down-regulate dlp2 expression (15, 25). Thus this situation in flies could potentially represent another case of FOXO-to-FOXO signaling.
Materials and Methods
Mutations.
Mutations were as follows: LGI, daf-16(mu86), a null allele (9); LGIII, daf-2(e1370); LGIV, ins-7(tm1907), ins-7(tm2001), ins-7(ok1573), ZK1251.1(tm1849), and ZK1251.3(tm1723).
Strains.
Strains were as follows: CE284, Pins-7::gfp (20); CF2362, Pins-7::gfp; daf-2(e1370); CF2340, Pins-7::gfp; daf-16(mu86); CF2266, muEx340[Pges-1::rfp + Pges-1::ins-7]; TJ356, Pdaf-16::daf-16::gfp (6); and CF1553, muIs84 (Psod-3::gfp) (14); CQ1: ins-7(tm1907) outcrossed three times with wild type (N2); CQ4: ins-7(tm2001) outcrossed three times with N2; CQ3, ins-7(ok1573) outcrossed three times with N2; FX1849: ZK1251.1(tm1849); FX1723: ZK1251.3(tm1723).
Molecular Biology and Generation of Transgenic Worms.
To express ins-7 specifically in the intestine (Pges-1::ins-7), we introduced Acc65I and EcoRI sites immediately 5′ of the first ATG and 3′ of the stop codon of the ins-7 gene, respectively, by PCR amplification of C. elegans genomic DNA with TTTGGTACCATGCCACCAATAATTTTGGTTTTC and TTTGAATTCTTAAG GACAGCACTGTTTTCGAATG primers. The Acc65I/EcoRI fragment was inserted into the Acc65I/EcoRI sites of Pmyo-3::rfp (gift from C. I. Bargmann, Rockefeller University, New York). A 2.5-kb upstream regulatory sequence of myo-3 was excised with XbaI and BamHI and blunt-ended, and a SnaB1 fragment from pNL213 (14) that has a 3.3-kb upstream sequence of intestinal ges-1 gene was introduced. Our Pges-1::ins-7 construct was not tagged; instead, we inferred its presence from a coinjected Pges-1::rfp construct. Red intestinal fluorescence likely indicates ins-7 expression, because red intestinal fluorescence coincided with green intestinal fluorescence in control experiments in which the same Pges-1::rfp construct was coinjected with a Pges-1::gfp construct (SI Fig. 6).
Standard techniques were used to generate transgenic animals (26). Pges-1::ins-7 was injected into N2 animals at 100 ng/μl with intestinal Pges-1::rfp at 100 ng/μl as a coinjection marker.
RNAi.
Bacterial-feeding RNAi experiments were carried out as described (27, 28). Control vector, daf-2, and daf-16 RNAi vectors were from Dillin et al. (28), and the ins-7 RNAi clone was from the Ahringer library (27). Each clone was verified by PCR and sequence analysis, and isopropyl β-d-thiogalactoside was added to increase induction. We also wrote a perlscript to identify possible secondary RNAi targets of ins-7 (see SI Text), but no matches met the 200-nt threshold for secondary targets, suggesting that the clone targets only ins-7.
Survival Analyses.
The first day of adulthood was defined as t = 0, and the log-rank (Mantel–Cox) method was used to test the null hypothesis (StatView 5.01, SAS Software), as described (29). Lifespan experiments were carried out at 20°C, and for all experiments, n > 60 for each sample.
GFP Assays.
Populations of live worms were scored for GFP by using a fluorescence dissecting microscope. The transgenic arrays carried by the animals we examined were extrachromosomal and therefore meiotically unstable. Because of this, a consistent fraction of the progeny of all GFP-expressing transgenic worms exhibited no GFP signal or Rol (coinjection marker) phenotype; these animals were discarded. The fraction of worms expressing intestinal GFP fluorescence and the standard error of the proportion (SEP) was determined. To avoid any artifacts due to prolonged fixation, all assays were performed on live nonanesthetized worms. n > 75 for all Pins-7::gfp assays.
Psod-3::gfp Assay.
The expression of Psod-3::gfp was assayed as described (14). Briefly, well fed 3-day-old adult animals grown at 20°C were mounted on 2% agarose slides (≈10 per slide). The expression of Psod-3::gfp in head muscles was assayed by using a combination of fluorescence and Nomarski microscopy.
Quantitative RT-PCR.
Purification and reverse transcription of RNA were carried out as described (30). Quantitative RT-PCR was performed in a 7300 Real Time PCR System (Applied Biosystems) and analyzed by using the Ct method (Applied Biosystems Prism 7700 Users Bulletin No. 2). mRNA levels of the actin gene, act-1, were used as controls for normalization.
Supplementary Material
Acknowledgments
We thank Malene Hansen, other Kenyon laboratory members, and Zemer Gitai (Princeton University, Princeton, NJ) for helpful discussions; Wendy Shaw and Jasmine Ashraf (Princeton University) for assistance; Mike Costa (Exelixis), Gary Ruvkun (Harvard Medical School, Boston, MA), Shohei Mitani (Tokyo Women's Medical University, Tokyo), the C. elegans Genetics Center for C. elegans strains; and Julie Ahringer (Cambridge University, Cambridge, U.K.) for RNAi clones. Z. Gitai and C.T.M. wrote the 22-nt scanning program. C.T.M. was a senior postdoctoral fellow of the Ellison Medical Foundation/American Federation for Aging Research. S.-J.L. is an Ellison Medical Foundation Fellow of the Life Sciences Research Foundation. C.K. is an American Cancer Society Research Professor and a founder of Elixir Pharmaceuticals. This work was supported by National Institutes of Health Grant RO1 AG11816. C.T.M. carried out the experiments showing that ins-7 is likely to be a DAF-2 agonist (Fig. 1 and SI Fig. 5) and analyzed the regulation of ins-7 (Fig. 2). S.-J.L. showed that ins-7 influences FOXO-to-FOXO signaling (Figs. 3 and 4 and SI Fig. 6; Table 1). C.T.M. and S.-J.L. both analyzed ins-18 (unpublished data and SI Fig. 7).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709613104/DC1.
References
- 1.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 2.Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herndon LA, Schmeissner PJ, Dudaronek JM, Brown PA, Listner KM, Sakano Y, Paupard MC, Hall DH, Driscoll M. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- 4.Apfeld J, Kenyon C. Cell. 1998;95:199–210. doi: 10.1016/s0092-8674(00)81751-1. [DOI] [PubMed] [Google Scholar]
- 5.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 6.Henderson ST, Johnson TE. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- 7.Lee RY, Hench J, Ruvkun G. Curr Biol. 2001;11:1950–1957. doi: 10.1016/s0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- 8.Lin K, Dorman JB, Rodan A, Kenyon C. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 9.Lin K, Hsin H, Libina N, Kenyon C. Nat Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- 10.Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 11.Kenyon C. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Tatar M, Bartke A, Antebi A. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- 13.Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 14.Libina N, Berman JR, Kenyon C. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- 15.Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- 16.Giannakou ME, Partridge L. Trends Cell Biol. 2004;14:408–412. doi: 10.1016/j.tcb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 17.Bluher M, Kahn BB, Kahn CR. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- 18.Honda S, Ishii N, Suzuki K, Matsuo M. J Gerontol. 1993;48:B57–B61. doi: 10.1093/geronj/48.2.b57. [DOI] [PubMed] [Google Scholar]
- 19.Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA. Nat Genet. 2006;38:251–257. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- 20.Pierce SB, Costa M, Wisotzkey R, Devadhar S, Homburger SA, Buchman AR, Ferguson KC, Heller J, Platt DM, Pasquinelli AA, et al. Genes Dev. 2001;15:672–686. doi: 10.1101/gad.867301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Apfeld J, Kenyon C. Nature. 1999;402:804–809. doi: 10.1038/45544. [DOI] [PubMed] [Google Scholar]
- 22.Li W, Kennedy SG, Ruvkun G. Genes Dev. 2003;17:844–858. doi: 10.1101/gad.1066503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kulkarni RN, Bruning JC, Winnay JN, Postic C, Magnuson MA, Kahn CR. Cell. 1999;96:329–339. doi: 10.1016/s0092-8674(00)80546-2. [DOI] [PubMed] [Google Scholar]
- 24.Ookuma S, Fukuda M, Nishida E. Curr Biol. 2003;13:427–431. doi: 10.1016/s0960-9822(03)00108-8. [DOI] [PubMed] [Google Scholar]
- 25.Wang MC, Bohmann D, Jasper H. Cell. 2005;121:115–125. doi: 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 26.Mello C, Fire A. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 27.Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 28.Dillin A, Crawford DK, Kenyon C. Science. 2002;298:830–834. doi: 10.1126/science.1074240. [DOI] [PubMed] [Google Scholar]
- 29.Lawless JF. Models and Methods for Lifetime Data. New York: Wiley; 1982. [Google Scholar]
- 30.Taubert S, Van Gilst MR, Hansen M, Yamamoto KR. Genes Dev. 2006;20:1137–1149. doi: 10.1101/gad.1395406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolkow CA, Kimura KD, Lee MS, Ruvkun G. Science. 2000;290:147–150. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- 32.McElwee J, Bubb K, Thomas JH. Aging Cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 33.Lee SS, Kennedy S, Tolonen AC, Ruvkun G. Science. 2003;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- 34.Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA. Nat Genet. 2006;38:251–257. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- 35.Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Curr Biol. 2002;12:1293–1300. doi: 10.1016/s0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.