Abstract
Amatoxins, the lethal constituents of poisonous mushrooms in the genus Amanita, are bicyclic octapeptides. Two genes in A. bisporigera, AMA1 and PHA1, directly encode α-amanitin, an amatoxin, and the related bicyclic heptapeptide phallacidin, a phallotoxin, indicating that these compounds are synthesized on ribosomes and not by nonribosomal peptide synthetases. α-Amanitin and phallacidin are synthesized as proproteins of 35 and 34 amino acids, respectively, from which they are predicted to be cleaved by a prolyl oligopeptidase. AMA1 and PHA1 are present in other toxic species of Amanita section Phalloidae but are absent from nontoxic species in other sections. The genomes of A. bisporigera and A. phalloides contain multiple sequences related to AMA1 and PHA1. The predicted protein products of this family of genes are characterized by a hypervariable “toxin” region capable of encoding a wide variety of peptides of 7–10 amino acids flanked by conserved sequences. Our results suggest that these fungi have a broad capacity to synthesize cyclic peptides on ribosomes.
Keywords: amanitin, cyclic peptide, phalloidin, phallotoxin, amatoxin
Mushrooms in the genus Amanita section Phalloideae account for >90% of all fatal mushroom poisonings (1). The human LD50 for α-amanitin (Fig. 1A) is ≈0.1 mg/kg, and one mature destroying angel (A. bisporigera, A. virosa, A. suballiacea, and allied species) (Fig. 2A) or death cap (A. phalloides) (Fig. 2B) can contain a fatal dose of 10–12 mg (2). Only the carpophores (fruiting bodies) contain high concentrations of the toxins. Like other ectomycorrhizal basidiomycetes, species of Amanita grow slowly and do not form carpophores in culture (3). There are ≈900–1,000 species of Amanita, but most do not produce amatoxins or phallotoxins, and some are edible (Fig. 2C) (4, 5).
Fig. 1.
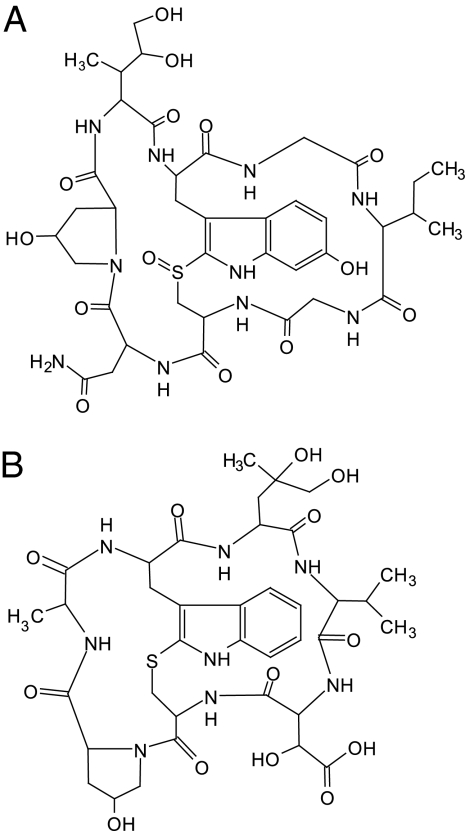
Structures of α-amanitin (A) and phallacidin (B). All of the amino acids have the l configuration except hydroxyAsp in phallacidin (Thr in phalloidin).
Fig. 2.
Fungi of the genus Amanita. (A) A. bisporigera (Oakland County, MI). (B) A. phalloides (Alameda County, CA). (C) Nondeadly species of Amanita. Shown from left to right are three specimens of A. gemmata, one specimen of A. muscaria, and two specimens of A. franchetii (Mendocino County, CA).
The mammalian toxicity of amatoxins is because of active cellular uptake followed by inhibition of RNA polymerase II (6–9). The typical symptoms of amatoxin poisoning are gastrointestinal distress beginning 6–12 h after ingestion, a remission phase lasting 12–24 h, and progressive loss of liver function culminating in death within 3–5 days. One of the few effective treatments is liver transplantation (10).
In addition to amatoxins, several members of Amanita section Phalloideae produce bicyclic heptapeptides called phallotoxins (Fig. 1B). Although structurally related to amatoxins, phallotoxins have a different mode of action, which is the stabilization of F-actin (11). Phallotoxins are poisonous when administered parenterally, but not orally because of poor absorption.
The biosynthetic origin of the Amanita toxins has been unknown. Because of the difficulty of working with Amanita fungi in culture, we took a genomic approach to identify genes involved in the biosynthesis of the amatoxins and phallotoxins.
Results and Discussion
The genome of A. bisporigera, an amatoxin- and phallotoxin-producing species native to North America (Fig. 2A), was shotgun-sequenced to approximately two times the coverage of the genome (≈70 MB total based on the known size of other homobasidiomycetes) (12) by a combination of automated Sanger sequencing and pyrosequencing (13). Because all known fungal cyclic peptides are biosynthesized by nonribosomal peptide synthetases (NRPSs) (14, 15), the genome survey sequences were first queried with known bacterial and fungal NRPSs. No evidence for any NRPS was found in A. bisporigera; the most closely related sequences were orthologs of aminoadipate reductase and acyl-CoA synthase, which are other members of the aminoacyl-adenylating superfamily (15).
We then searched the A. bisporigera genome for DNA encoding amanitins' amino acid sequences. Simplifed to the unmodified 20 proteogenic amino acids (i.e., ignoring the hydroxylations and Trp-Cys cross-bridge) (Fig. 1), the sequence of the amatoxins is a cyclic permutation of either IWGIGCNP (α- and γ-amanitins) or IWGIGCDP (β- and ε-amanitins). Nucleotide sequences that could encode the amino acid sequence of α-amanitin were found in the genome of A. bisporigera. This sequence is not present in any protein or gene in the GenBank database, therefore it is not likely to be present in A. bisporigera by chance. Inverse PCR by using the restriction enzyme PvuI resulted in the isolation of a 2.5-kb fragment of flanking genomic DNA. An RNA blot probed with this DNA indicated that this region of the genome is transcribed into an mRNA of <400 nt (data not shown). PCR primers based on the genomic sequence were used to amplify a cDNA of ≈380 bp by 3′ and 5′ rapid amplification of cDNA ends (RACE). Comparison of the cloned, polyadenylated cDNA to the genomic sequence indicated that the gene, AMA1, has three introns with conventional GT/AG intron borders. Two of the introns (53 and 59 nt in length) are in the 3′ untranslated region, and one intron (58 nt) interrupts the fourth from the last codon (Fig. 3A). The presence of these features indicates that AMA1 constitutes a true transcribed and processed gene. Assuming that translation starts at the first ATG downstream of the transcriptional start site, AMA1 encodes a proprotein of 35 amino acids (Fig. 3A).
Fig. 3.
Nucleotide sequences of cDNAs for AMA1 and PHA1. (A) AMA1. The sequence of α-amanitin is underlined. Carets indicate the positions of the three introns. (B) PHA1. The sequence of phallacidin is underlined. Carets indicate the positions of the three introns.
A genomic survey sequence of A. bisporigera also predicted the peptide AWLVDCP, which matches phallacidin, one of the major phallotoxins (Fig. 1B). Inverse PCR using PvuI and SacI was used to isolate genomic fragments of 1.6 and 1.9 kb, respectively, covering the PHA1 gene. Two different classes of sequences were found, which were identical in the region of phallacidin but diverged ≈135 nt upstream. This finding indicates that A. bisporigera has at least two copies of the PHA1 gene, both of which could encode phallacidin. A cDNA for PHA1 was isolated by using 3′ and 5′ RACE. Like AMA1, the cDNA for PHA1 also has three introns (57, 70, and 51 nt in length) in approximately the same positions as the introns in AMA1. The proprotein of PHA1 is 34 amino acids (Fig. 3B).
AMA1 and PHA1 and their translation products are similar in overall size and sequence (Fig. 4). The translated regions upstream of the toxin sequences have 28 of 30 nt in common (93%), the regions downstream have 40 of 48 nt in common (83%), but the toxin regions have only 11 of 24 nt in common (46%). Thus, the proproteins of α-amanitin and phallacidin are composed of two domains, a variable toxin region flanked by conserved regions (Fig. 4).
Fig. 4.
Alignment of the cDNA nucleotide (A) and predicted amino acid sequences (B) of the coding regions of AMA1 and PHA1. The mature toxin sequences are underlined.
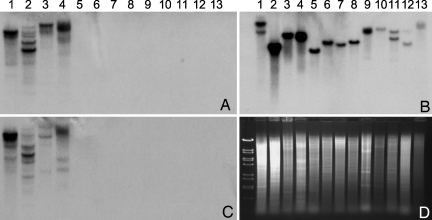
Many secondary metabolites are limited in their taxonomic distribution, and most species of Amanita do not make amatoxins or phallotoxins. To test whether the lack of toxin production among other species of Amanita were because of absence of the encoding genes, a blot of genomic DNA from 12 species of Amanita was hybridized with AMA1 and PHA1. The species include four from section Phalloideae (this section contains all of the species that make amatoxins and phallotoxins), three from section Validae (the sister group to section Phalloideae), two from section Amanita, one from section Caesarea, and two from section Vaginatae (4, 5). All mushrooms were tested and confirmed by HPLC for the expected presence or absence of amatoxins and phallotoxins. All of the tested species that synthesize amatoxins and phallotoxins, but none of the nonproducers, hybridize to AMA1 and PHA1 (Fig. 5). This finding is consistent with AMA1 and PHA1 being responsible for amanitin and phallacidin biosynthesis and provides a molecular explanation for why Amanita species outside of section Phalloideae are not deadly poisonous. (Some of the Amanita species that do not make amatoxins or phallotoxins are edible, but others make different toxic compounds.)
Fig. 5.
DNA blots of different species of Amanita. (A) Probed with AMA1 cDNA. (B) Probed with PHA1 cDNA. (C) Probed with a fragment of the β-tubulin gene isolated from A. bisporigera (see SI Text). (D) Ethidium-stained gel showing relative lane loading. Markers are λ phage DNA cut with BstEII. Species and provenances are as follows: lane 1, A. aff. suballiacea (Ingham County, MI); lane 2, A. bisporigera (Ingham County); lane 3, A. phalloides (Alameda County, CA); lane 4, A. ocreata (Sonoma County, CA); lane 5, A. novinupta (Sonoma County); lane 6, A. franchetii (Mendocino County, CA); lane 7, A. porphyria (Sonoma County); lane 8, a second isolate of A. franchetii (Sonoma County); lane 9, A. muscaria (Monterey County, CA); lane 10, A. gemmata (Mendocino County); lane 11, A. hemibapha (Mendocino County); lane 12, A. velosa (Napa County, CA); and lane 13, Amanital section Vaginatae (Mendocino County). Mushrooms represent sections Phalloideae (1–4), Validae (5–8), Amanita (9 and 10), Caesareae (11), and Vaginatae (12 and 13). Four separate gels were run; the lanes are in the same order on each gel, and approximately the same amount of DNA was loaded per lane. A and B are to the same scale, and C and D are to the same scale.
The complex hybridization patterns shown in Fig. 5 indicate that AMA1 and PHA1 are members of gene families. Therefore, the conserved upstream and downstream amino acid sequences of AMA1 and PHA1 were used as queries to search for additional related sequences in the A. bisporigera genome. We thereby found at least 13 new, related complete or almost complete sequences (Fig. 6A) and another 10–15 sequences missing one end or the other (data not shown). All of these new sequences have an upstream conserved consensus sequence MSDINTARLP (MSDIN, R, and P are invariant) and a downstream conserved consensus sequence CVGDDV (the first D is invariant). The putative toxin regions, which start immediately downstream of the invariant Pro residue and end after an invariant Pro residue, are hypervariable compared with the upstream and downstream sequences. The hypervariable regions contain 7–10 amino acids, and all 20 proteogenic amino acids are represented at least once.
Fig. 6.
Sequences related to AMA1 and PHA1. (A) Related, predicted amino acid sequences identified in the A. bisporigera genome. (B) PCR products amplified from A. phalloides and A. ocreata (phalloidin) with degenerate primers based on the conserved sequences of AMA1 and PHA1. Spaces have been inserted after some of the toxin regions (underlined) to emphasize the conservation of the downstream sequences. Asterisks indicate stop codons.
To detect related genes in A. phalloides, which worldwide accounts for the majority of fatal mushroom poisonings, degenerate PCR primers were designed against the conserved upstream and downstream sequences of AMA1 and PHA1. The predicted translations of four amplicons from A. phalloides and one from A. ocreata are shown in Fig. 6B. One of them (IWGIGCDP) matches the amino acid sequence of β-amanitin, one matches phalloidin (AWLATCP), and the other two predict novel peptides.
The results in Fig. 6 suggest that species of Amanita section Phalloideae have the capacity to synthesize small, cyclic peptides in addition to amatoxins and phallotoxins. In fact, A. phalloides is known to produce other cyclic peptides, including CyA-A, CyA-B, CyA-C, CyA-D, and antamanide, which have the structures cyclo(GVAFFP), cyclo(SFFFPIP), cyclo(MLGFLVLP), cyclo(MLGFLPLP), and cyclo(FFVPPAFFPP), respectively (2, 16–18). None of these amino acid sequences were found in the genome survey sequences of A. bisporigera, but FFQPPEFRPP (Fig. 6A) is 70% identical to antamanide (18).
Small, modified, and biologically active peptides were previously identified from bacteria and several animals, including arachnids, snakes, cone snails, and amphibian skin (19–21). Like the Amanita toxins, the animal peptides are synthesized as precursor proteins and often undergo posttranslational modifications, including, like the Amanita toxins, hydroxylation and epimerization (22–24). Both the conotoxin and the Amanita toxin genes are characterized by the presence of conserved and hypervariable regions, resulting in the capacity to synthesize a large number of peptides on the same fundamental biosynthetic scaffold (Fig. 6) (25).
The Amanita toxins differ from these other small peptides in several key aspects. First, the animal peptides are not cyclized by peptide bonds, but acquire their essential rigidity by extensive disulfide bonds. Second, although ribosomally synthesized cyclic peptides have been described for bacteria, plants, and animals (e.g., the cyclotides and microcin J25) (26, 27), to the best of our knowledge, all previously known fungal cyclic peptides are synthesized by nonribosomal peptide synthetases (14, 15). Third, the Amanita toxins are not secreted (3), and, consistent with this fact, they lack predicted signal peptides (Figs. 3–5). Fourth, whereas the animal peptides are processed from their respective proproteins by proteases that recognize basic amino acid residues (Arg or Lys) (19, 24), we predict that the toxins of Amanita are cleaved from their proproteins by a protease that hydrolyzes peptide bonds specifically at Pro. All of the known Amanita cyclic peptides contain Pro, the last amino acid in the upstream conserved region is always Pro, and the predicted toxin sequences all have Pro as the last amino acid (Figs. 4 and 6).
Based on the properties of the known proline-specific peptidases (28, 29), the prolyl oligopeptidase family (POP) (EC 3.4.21.26) is the most promising to be involved in the processing of the proproteins of the Amanita toxins. We identified sequences related to human POP (GenBank accession no. NP_002717) in the genome survey sequences of A. bisporigera [see supporting information (SI) Text]. Orthologs of human POP also were found in every other basidiomycete for which whole genome sequences are available (Laccaria bicolor, Coprinus cinereus, Phanerochaete chrysosporium, Ustilago maydis, Sporobolomyces roseus, Puccinia graminis, and Cryptococcus neoformans) (see SI Text). A POP has been characterized from the mushroom Lyophyllum cinerascens (30). In contrast, orthologs of human POP are rare or nonexistent in fungi outside of the basidiomycetes. BLASTP (default parameters) identified no orthologs of human POP with a score >53 and E value <10−6 in any fungus outside of the basidiomycetes, except perhaps in the ascomycete Setosphaeria nodorum (SNOG11288; score = 166; E value = 3 × 10−40). Thus, it appears that at least one component of the biochemical machinery necessary for the biosynthesis of the Amanita toxins is both widespread in, and restricted to, the basidiomycetes.
The results presented here indicate that species of Amanita section Phalloidae synthesize their notoriously toxic cyclic peptides on ribosomes. Furthermore, these fungi have evolved a unique mechanism of combinatorial biosynthesis that endows them with the ability to biosynthesize a multitude of cyclic peptides. Further elucidation of the biosynthetic pathway of Amanita toxin biosynthesis could take advantage of the tractability of some basidiomycete fungi such as C. cinereus (31).
Materials and Methods
Mushrooms were harvested from the wild in 2002, 2006, and 2007; frozen at −80°C; and lyophilized. DNA was extracted from lyophilized fruiting bodies or cultures by using cetyltrimethyl-ammonium bromide, phenol, and chloroform (32). RNA was extracted by using TRIzol (Invitrogen) (33).
PCR products were purified by using Wizard SV Gel and PCR Clean-Up System (Promega) and were cloned into TOPO pCR 4 (Invitrogen) for sequencing. For 3′ RACE, initial and nested primers from GeneRacer (Invitrogen) were used, and gene-specific primers were derived from the genomic sequence. Primer sequences may be found in SI Text.
Probe labeling, DNA blotting, and filter hybridization followed standard protocols (34, 35). DNA for blotting was cut with PstI and electrophoresed in 0.7% agarose. Hybridizations were performed overnight at 65°C in 4× SET, 0.1% sodium pyrophosphate, 0.2% SDS, 10% dextran sulfate, and 625 μg/ml heparin. SET (20×) is 3 M NaCl, 0.6 M Tris, and 0.04 M EDTA (pH 7.4). A 551-bp fragment of the A. bisporigera β-tubulin gene used as a control probe on DNA blots was amplified by PCR.
Variability in toxin content is known within species of Amanita (36, 37). All fungi analyzed for the presence of AMA1 and PHA1 (Fig. 5) were analyzed for amatoxins and phallotoxins by established HPLC methods (32, 38). Standards of α-amanitin, β-amanitin, phalloidin, and phallacidin were purchased from Sigma–Aldrich.
Supplementary Material
Acknowledgments
This article is dedicated to the memory of Hans Kende (1937–2006), whose inspiration and support made this work possible. We thank Darvin DeShazer (St. Vincent High School, Petaluma, CA), Anne Pringle (Harvard University, Cambridge, MA), Daphne O'Regan (Michigan State University College of Law), Brian O'Regan (University College, London, U.K.), Suzanne Kiihne, Ursula Schulz (Cheese Board Collective, Berkeley, CA), and Christopher Walton for supplying mushroom specimens, and the Michigan State University Research Technical Support Facility for DNA sequencing. This work was supported by a grant from the U.S. Department of Energy, Division of Energy Biosciences, to the Plant Research Laboratory, and by a Strategic Partnership Grant from the Michigan State University Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU196139–EU196158).
This article contains supporting information online at www.pnas.org/cgi/content/full/0707340104/DC1.
References
- 1.Bresinsky A, Besl H. A Colour Atlas of Poisonous Fungi: A Handbook for Pharmacists, Doctors and Biologists. Würzburg, Germany: Wolfe; 1990. [Google Scholar]
- 2.Wieland T. Peptides of Poisonous Amanita Mushrooms. New York: Springer; 1986. [Google Scholar]
- 3.Zhang P, Chen Z, Hu J, Wei B, Zhang Z, Hu W. FEMS Microbiol Lett. 2005;252:223–228. doi: 10.1016/j.femsle.2005.08.049. [DOI] [PubMed] [Google Scholar]
- 4.Tulloss RE. Boll Gruppo Micologico G Bresadola. 2000;43:13–21. [Google Scholar]
- 5.Weiβ M, Yang Z-L, Oberwinkler F. Can J Bot. 1998;76:1170–1179. [Google Scholar]
- 6.Bushnell DA, Cramer P, Kornberg RD. Proc Natl Acad Sci USA. 2002;99:1218–1222. doi: 10.1073/pnas.251664698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lengsfeld AM, Low I, Wieland T, Dancker P, Hasselbach W. Proc Natl Acad Sci USA. 1974;71:2803–2807. doi: 10.1073/pnas.71.7.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kröncke KD, Fricker G, Meier PJ, Gerok W, Wieland T, Kurz G. J Biol Chem. 1986;261:2562–2567. [PubMed] [Google Scholar]
- 9.Letschert K, Faulstich H, Keller D, Keppler D. Toxicol Sci. 2006;91:140–149. doi: 10.1093/toxsci/kfj141. [DOI] [PubMed] [Google Scholar]
- 10.Enjalbert F, Rapior S, Nouguier-Soule J, Guillon S, Amouroux N, Cabot C. J Toxicol Clin Toxicol. 2002;40:715–757. doi: 10.1081/clt-120014646. [DOI] [PubMed] [Google Scholar]
- 11.Bamburg JR. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- 12.Le Quéré A, Johansson T, Tunlid A. Fung Genet Biol. 2002;36:234–241. doi: 10.1016/s1087-1845(02)00024-5. [DOI] [PubMed] [Google Scholar]
- 13.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walton JD, Panaccione DG, Hallen HE. In: Advances in Fungal Biotechnology for Industry, Agriculture, and Medicine. Tkacz JS, Lange L, editors. New York: Kluwer; 2004. pp. 127–162. [Google Scholar]
- 15.Finking R, Marahiel MA. Annu Rev Microbiol. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 16.Gauhe A, Wieland T. Justus Liebigs Ann Chem. 1977:859–868. [Google Scholar]
- 17.Chiang CC, Karle IL, Wieland T. Int J Peptide Protein Res. 1982;20:414–420. doi: 10.1111/j.1399-3011.1982.tb03061.x. [DOI] [PubMed] [Google Scholar]
- 18.Wieland T, Lüben G, Otttenheym H, Faesel J, deVries JX, Konz W, Prox A, Schmid J. Angew Chem Int Ed Engl. 1968;7:204–208. [Google Scholar]
- 19.Escoubas P. Mol Divers. 2006;10:545–554. doi: 10.1007/s11030-006-9050-4. [DOI] [PubMed] [Google Scholar]
- 20.Olivera BM. J Biol Chem. 2006;281:31173–31177. doi: 10.1074/jbc.R600020200. [DOI] [PubMed] [Google Scholar]
- 21.Simmaco M, Mignogna G, Barra D. Biopolymers. 1998;47:435–450. doi: 10.1002/(SICI)1097-0282(1998)47:6<435::AID-BIP3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 22.Buczek O, Bulaj G, Olivera BM. Cell Mol Life Sci. 2005;62:3067–3079. doi: 10.1007/s00018-005-5283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shikata Y, Watanabe T, Teramoto T, Inoue A, Kawakami Y, Nishizawa Y, Katayama K, Kuwada M. J Biol Chem. 1995;270:16719–16723. doi: 10.1074/jbc.270.28.16719. [DOI] [PubMed] [Google Scholar]
- 24.Richter K, Egger R, Negri L, Corsi R, Severini C, Kreil G. Proc Natl Acad Sci USA. 1990;87:4836–4839. doi: 10.1073/pnas.87.12.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodward SR, Cruz LJ, Olivera BM, Hillyard DR. EMBO J. 1990;9:1015–1020. doi: 10.1002/j.1460-2075.1990.tb08204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Craik DJ, Cemazar M, Daly NL. Curr Opin Drug Discov Devel. 2007;10:176–184. [PubMed] [Google Scholar]
- 27.Rosengren KJ, Clark RJ, Daly NL, Goransson U, Jones A, Craik DJ. J Am Chem Soc. 2003;125:12464–12474. doi: 10.1021/ja0367703. [DOI] [PubMed] [Google Scholar]
- 28.Cunningham DF, O'Connor B. Biochim Biophys Acta. 1997;1343:160–186. doi: 10.1016/s0167-4838(97)00134-9. [DOI] [PubMed] [Google Scholar]
- 29.Polgár L. Cell Mol Life Sci. 2002;59:349–362. doi: 10.1007/s00018-002-8427-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshimoto T, Sattar AK, Hirose W, Tsuru D. J Biochem. 1988;104:622–627. doi: 10.1093/oxfordjournals.jbchem.a122522. [DOI] [PubMed] [Google Scholar]
- 31.Kues U. Microbiol Mol Biol Rev. 2000;64:316–353. doi: 10.1128/mmbr.64.2.316-353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hallen HE, Watling R, Adams GC. Mycol Res. 2003;107:969–979. doi: 10.1017/s0953756203008190. [DOI] [PubMed] [Google Scholar]
- 33.Hallen HE, Huebner M, Shiu S-H, Güldener U, Trail F. Fung Genet Biol. 2007;44:1146–1156. doi: 10.1016/j.fgb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1982. [Google Scholar]
- 35.Singh L, Jones KW. Nucleic Acids Res. 1984;12:5627–5638. doi: 10.1093/nar/12.14.5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beutler JA, der Marderosian AH. J Nat Prod. 1981;44:422–431. [Google Scholar]
- 37.Tyler VE, Jr, Benedict RG, Brady LR, Robbers JE. J Pharm Sci. 1966;55:590–593. doi: 10.1002/jps.2600550612. [DOI] [PubMed] [Google Scholar]
- 38.Enjalbert F, Gallion C, Jehl F, Monteil H, Faulstich H. J Chromatogr. 1992;598:227–236. doi: 10.1016/0021-9673(92)85052-u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.