Abstract
Iron is essential for most living organisms and is often the major limiting nutrient for normal growth. Plants induce iron utilization systems under conditions of low iron availability, but the molecular mechanisms of gene regulation under iron deficiency remain largely unknown. We identified the rice transcription factor IDEF1, which specifically binds the iron deficiency-responsive cis-acting element IDE1. IDEF1 belongs to an uncharacterized branch of the plant-specific transcription factor family ABI3/VP1 and exhibits the sequence recognition property of efficiently binding to the CATGC sequence within IDE1. IDEF1 transcripts are constitutively present in rice roots and leaves. Transgenic tobacco plants expressing IDEF1 under the control of the constitutive cauliflower mosaic virus 35S promoter transactivate IDE1-mediated expression only in iron-deficient roots. Transgenic rice plants expressing an introduced IDEF1 exhibit substantial tolerance to iron deficiency in both hydroponic culture and calcareous soil. IDEF1 overexpression leads to the enhanced expression of the iron deficiency-induced transcription factor gene OsIRO2, suggesting the presence of a sequential gene regulatory network. These findings reveal cis element/trans factor interactions that are functionally linked to the iron deficiency response. Manipulation of IDEF1 also provides another approach for producing crops tolerant of iron deficiency to enhance food and biomass production in calcareous soils.
Keywords: ABI3/VP1, iron deficiency-responsive elements, mugineic acid family phytosiderophores, transgenic rice
Most living organisms require iron for growth and reproduction, and the iron absorbed by plants constitutes a major iron source for animals and humans. Although abundant in mineral soils, iron is sparingly soluble under aerobic conditions at high pH. Consequently, in calcareous soils, which constitute ≈30% of the world's cultivated soils, plants often exhibit iron deficiency symptoms manifested as chlorosis (yellowing caused by chlorophyll deficiency), reducing crop yield and quality (1). Higher plants use two major iron uptake strategies under conditions of low iron supply: reduction (Strategy I) and chelation (Strategy II) (2). The Strategy II mechanism is specific to graminaceous plants and is mediated by natural iron chelators, the mugineic acid family phytosiderophores (MAs) (3). Extensive physiological, biochemical, and molecular studies have identified the biosynthetic pathway of MAs and the genes encoding the biosynthetic enzymes (4–9). The expression of these biosynthetic genes is coordinately up-regulated in response to iron deficiency (6, 8, 9), but the molecular mechanisms regulating these iron deficiency-induced genes are poorly understood.
Recent studies have demonstrated that a rice iron deficiency-induced basic helix—loop–helix (bHLH) transcription factor, OsIRO2, regulates the Strategy II-based iron deficiency response by inducing the related genes (10, 11). The core sequence to which OsIRO2 binds (CACGTGG) is overrepresented in iron deficiency-inducible gene promoters in rice (10), but its actual function in a given promoter has not been confirmed. In nongraminaceous plants, the involvement of several bHLH transcription factors in the iron deficiency response, including tomato FER and the Arabidopsis FIT (formerly FIT1/FRU/AtbHLH29), AtbHLH38, and AtbHLH39, has been reported (12–15), but their functional cis sequences have not been determined.
We previously analyzed the promoter region of the barley iron deficiency-inducible IDS2 gene using transgenic tobacco plants, identifying the iron deficiency-responsive cis-acting elements IDE1 and IDE2 (iron deficiency-responsive elements 1 and 2) (16). IDE1 and IDE2 synergistically induce iron deficiency-specific gene expression in tobacco roots and in rice roots and leaves (16, 17). The promoter regions of many iron deficiency-inducible genes in barley, rice, and Arabidopsis possess IDE-like sequences (8, 16). In addition, a tobacco ABC transporter gene containing an IDE1-like sequence in its promoter region, NtPDR3, was found to be inducible by iron deficiency (18). These facts suggest that gene regulation mechanisms involving IDEs not only are conserved among graminaceous (Strategy II) plants but also are functional in nongraminaceous (Strategy I) plant species. No transcription factors that interact with IDEs have been reported to date. In the present study, we identified a rice transcription factor, IDEF1, which specifically binds to IDE1. We provide evidence that IDEF1 functions as a key component regulating the response to and tolerance of iron deficiency.
Results and Discussion
Identification of IDEF1 as a Transcription Factor Recognizing the CATGC Sequence Within IDE1.
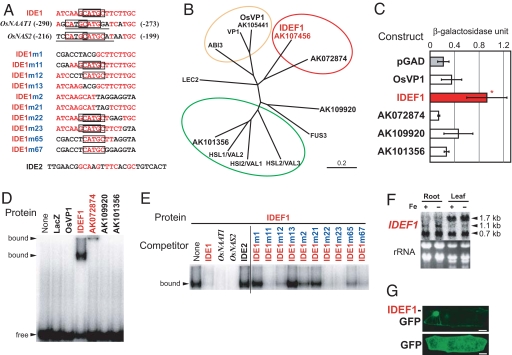
To identify candidates for IDE1-binding transcription factors, we first searched for known recognition sequences of transcription factors in IDE1 and IDE1-like sequences. IDE1 was found to possess a CATGC sequence similar to the Sph motif (TCCATGCAT)/RY element (CATGCA), which is recognized by the plant-specific B3 DNA-binding domain of the ABI3/VP1 family of transcription factors (19–21). Furthermore, the IDE1-like sequences present within proximal regions of the OsNAAT1 and OsNAS2 promoters (8, 16) possess a canonical Sph motif/RY element (Fig. 1A). ABI3/VP1-family transcription factors transmit the abscisic acid signal and transactivate various genes during seed maturation (19, 21–24). In rice, OsVP1 is a functional ortholog of the maize VIVIPAROUS 1 (VP1) (22) and is the only characterized member of the ABI3/VP1 family. A database search revealed four genes homologous to the B3 domain of OsVP1 (AK107456, AK072874, AK109920, and AK101356; Fig. 1B). These four genes showed no pronounced homology to OsVP1 outside of the B3 domain. A yeast assay system and an EMSA were used to confirm the binding activity of rice ABI3/VP1 members to IDE1 in vivo and in vitro. In yeast cells carrying the lacZ gene under the control of three tandem repeats of IDE1, only AK107456 induced substantial LacZ activity (Fig. 1C); this we designated as IDE-binding factor 1 (IDEF1). Similar results were obtained by using two tandem repeats of IDE2 and IDE1 fused upstream of lacZ (data not shown). Consistent with the yeast results, EMSA showed that IDEF1 specifically binds to IDE1 and Sph/RY (Fig. 1 D and E). The AK072874 protein also bound specifically to IDE1 and Sph/RY to form an unmobilized band on the plate [Fig. 1D; supporting information (SI) Fig. 5], whereas OsVP1, AK109920, and AK101356 exhibited no specific binding to IDE1 (Fig. 1D). The precise recognition sequence of IDEF1 and AK072874 was determined by competition experiments using mutated IDE1 sequences (Fig. 1 A and E; SI Fig. 5). IDEF1 and AK072874 specifically recognized the CATGC sequence present within IDE1, which is shorter than the previously reported minimal recognition sequence (CATGCA) of ABI3/VP1 transcription factors (20, 21). We further examined the binding activity of IDEF1 to several IDE1-like sequences present within previously determined functional regions for the iron deficiency response in the HvNAS1 and IDS3 promoters (25, 26), as well as those within the proximal region of the iron deficiency-inducible HvNAAT-A gene. IDEF1 efficiently bound to CATGC-containing IDE1-like sequences but not to IDE1-like sequences lacking a CATGC sequence (SI Fig. 5).
Fig. 1.
Identification and characterization of IDEF1. (A) The IDE1, IDE1-like, and IDE2 sequences used for EMSA. Red letters, bases identical to IDE1; underlined, conserved Sph/RY element sequences; boxes, the determined IDEF1-binding sequences (CATGC); numerals in parentheses, positions from the translation start sites. (B) Phylogenic tree of the ABI3/VP1-family transcription factors. Subgroups deduced from published reports (19–24) and from the present study are indicated by circles: red, IDE1/Sph/RY-binding factors; orange, Sph/RY-binding factors transmitting the abscisic acid signal during seed maturation; green, repressors of seed maturation-related genes during germination. (C) Binding assay of yeast GAL4 activation domain-fused rice ABI3/VP1 family members for IDE1 binding in yeast carrying IDE1 × 3-lacZ. Shown are β-galactosidase activity units (means ± SD; n = 5) and significant differences as compared with a vector control (pGAD) analyzed by using a t test (*, P < 0.05). (D and E) EMSA of rice ABI3/VP1 family members for IDE1 binding. (D) An IDE1 probe was incubated with 100 ng each of ABI3/VP1-family proteins fused to maltose-binding protein (MBP) or the LacZ-MBP fusion protein (LacZ) as a control. (E) Competition experiments were carried out by adding a 30-fold excess of unlabeled competitors carrying the sequences shown in A. (F) Northern blot analysis of IDEF1 in rice roots and leaves grown under iron sufficiency (+) or deficiency (−) for 14 days. Each lane was loaded with 10 μg of total RNA. Estimated transcript lengths are indicated. (G) Subcellular localization of IDEF1 in onion epidermal cells. IDEF1 fused to GFP or GFP alone was transiently expressed in onion epidermal cells and observed by confocal microscopy. (Scale bars: 50 μm.)
Northern blot analysis was conducted to detect the expression of the ABI3/VP1 family of genes in rice roots and leaves during the vegetative stage. The IDEF1 transcript was constitutively expressed in roots and leaves, with no obvious regulation by iron deficiency (Fig. 1F). Several bands were detected. Because estimated length of the longest transcript (≈1.7 kb) is in agreement with the length of AK107456 cDNA, this transcript, which was abundant in leaves, presumably corresponds to full-length IDEF1. The other shorter transcripts might correspond to partial IDEF1 sequences. OsVP1, AK109920, and AK101356 were also constitutively expressed in roots and leaves, but AK072874 expression was not detected in these organs (data not shown). These results are in accordance with our previous microarray experiment using rice 22k array slides (10, 11, 27), which also indicated that IDEF1, in addition to OsVP1, is clearly expressed in germinating seeds (data not shown). Because of the substantial expression of IDEF1 in vegetative organs, in contrast to little expression of AK072874, which constitutes the subgroup of IDE1/Sph/RY-binding factors with IDEF1 (Fig. 1 B, D, and E; SI Fig. 5), we focused on further characterization of IDEF1 in relation to plant iron-deficiency response.
A transiently expressed IDEF1-GFP fusion localized to the nucleus in onion epidermal cells (Fig. 1G). A database search of ESTs revealed the presence of IDEF1 homologs in several graminaceous species, but no obvious homologs belonging to the IDEF1 subgroup were found in nongraminaceous plants (data not shown).
Transactivation Analysis of IDEF1 in Tobacco Plants.
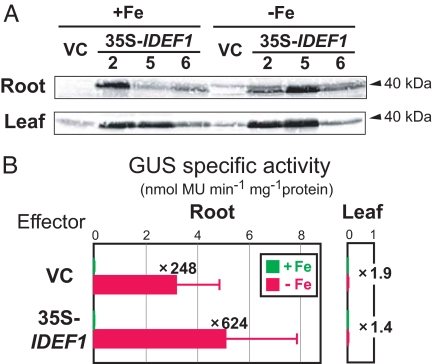
To characterize the in-planta function of IDEF1, we first produced transgenic tobacco plants containing two copies of IDE1 fused to the β-glucuronidase (GUS) gene as a reporter, and then introduced either the IDEF1 gene under the control of the constitutive 35S promoter (35S-IDEF1) or a vector control (VC) as an effector. 35S-IDEF1 tobacco plants had slightly higher chlorophyll contents under iron deficiency than VC plants (SI Fig. 6) but showed no other visible phenotypic differences. Constitutive expression of the IDEF1 transgene was detected in roots and leaves of 35S-IDEF1 plants (Fig. 2A). These transformants did not show substantial GUS activity in iron-sufficient roots or iron-sufficient or -deficient leaves (Fig. 2B). In iron-deficient roots, however, strong GUS activity driven by the duplicated IDE1 was observed in VC plants and was even more evident in 35S-IDEF1 plants (Fig. 2B). This iron-deficiency-induced and root-specific transactivation of GUS expression, despite the constitutive expression of the IDEF1 transgene, suggests a requirement for other factors for IDE-based activation in planta. These results also demonstrate that a duplicated IDE1 confers iron deficiency-induced and root-specific expression in native tobacco, in a fashion similar to the combination of IDE1 and IDE2 (16). Although we found no IDEF1 homologs in tobacco EST databases, Western blot analysis in VC plants detected weak bands at sizes similar to that of IDEF1 (Fig. 2A), suggesting the possible presence of IDEF1 homologs in tobacco. Alternatively, tobacco might possess other IDE1-binding factors distinct from IDEF1.
Fig. 2.
In-planta characterization of IDEF1 using transgenic tobacco. Double transformants were generated by introducing IDE1-IDE1-GUS as a reporter and 35S-IDEF1 or the vector control (VC) as an effector. (A) Western blot analysis of IDEF1 in T1 double transformants (VC line 19 and 35S-IDEF1 lines 2, 5, and 6); 40 μg of total protein were loaded per lane. Bands about the size of the estimated full-length IDEF1 (40 kDa) are indicated. (B) GUS activity in roots (Left) and leaves (Right) of T1 double transformants (12 and 10 independent lines containing 35S-IDEF1 and VC, respectively) grown under iron sufficiency (+Fe; green) or deficiency (−Fe; red) is shown as the mean ± SD. The ratio of GUS activity in iron-deficient and -sufficient roots is shown. Although no significant differences were detected between 35S-IDEF1 and VC by using a nonparametric Mann–Whitney test (P < 0.05), a similar higher induction in iron-deficient 35S-IDEF1 roots was observed in another set of experiments (data not shown).
The Induced Expression of IDEF1 in Rice Confers Iron-Deficiency Tolerance Through Gene Regulation.
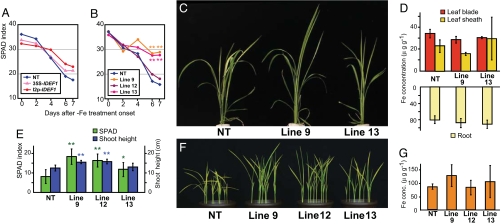
We introduced the IDEF1 gene into rice under the control of the 35S promoter (35S-IDEF1) or the iron deficiency-inducible IDS2 promoter (I2p-IDEF1) to investigate the function of IDEF1 in this crop. T1 transformants and nontransformants (NT) were grown hydroponically to determine the response of the plants to iron deficiency. With an adequate iron supply, the 35S-IDEF1 transformants showed substantially retarded early growth (SI Fig. 7), whereas the I2p-IDEF1 transformants grew similarly to the NT. With iron deficiency, the decline in leaf chlorophyll was slower in many 35S- and I2p-IDEF1 lines than in NT plants (Fig. 3A). Time-course observations of three representative I2p-IDEF1 lines (lines 9, 12, and 13) during iron-deficiency treatment revealed substantial tolerance of iron deficiency (Fig. 3 B and C). The I2p-IDEF1 plants grown in an iron-deficient hydroponic culture contained iron concentrations similar to those in NT plants in leaf blades, leaf sheaths, and roots (Fig. 3D). These transformants also grew better when germinated on a calcareous soil without additional micronutrient fertilizer, as confirmed by higher leaf chlorophyll contents and greater shoot lengths than in NT seedlings (Fig. 3 E and F). Aerial parts of I2p-IDEF1 seedlings grown in calcareous soil contained similar concentrations of iron to those of NT plants (Fig. 3G). These results indicate that the induced expression of IDEF1 in rice plants confers tolerance of iron deficiency, most likely through improved iron utilization within the plant body rather than enhanced iron uptake from the rhizosphere.
Fig. 3.
Tolerance of iron deficiency conferred by the induced expression of IDEF1 in rice. (A–D) Plants subjected to iron-deficiency treatment in hydroponic culture. (A) Mean chlorophyll contents (SPAD index) of the largest leaves of eight independent lines of 35S- and I2p-IDEF1 transformants and three NT plants. (B) SPAD index of the youngest leaves of I2p-IDEF1 transformants (lines 9, 12, and 13) and NT, and significant differences against NT analyzed by using a t test (**, P < 0.01). n = 3–8. (C) Iron deficiency-tolerant phenotype of I2p-IDEF1 transformants (lines 9 and 13) after 5 days of treatment. (D) Iron concentrations in leaf blades, leaf sheaths, and roots of I2p-IDEF1 transformants (lines 9 and 13) and NT after 12 days of treatment, expressed as microgram per gram dry weight; n = 3. (E–G) Seedlings germinated in a calcareous soil. (E) SPAD index of the largest leaves and shoot height of I2p-IDEF1 transformants (lines 9, 12, and 13) and NT 17 days after sowing. Shown are means ± SD (n = 12–20) and significant differences from NT, analyzed by using a t test (*, P < 0.05; **, P < 0.01). (F) Iron deficiency-tolerant phenotype of I2p-IDEF1 transformants (lines 9, 12, and 13) 17 days after sowing. (G) Iron concentrations in shoots of I2p-IDEF1 transformants (lines 9, 12, and 13) and NT 25 days after sowing, expressed as microgram per gram dry weight; n = 3.
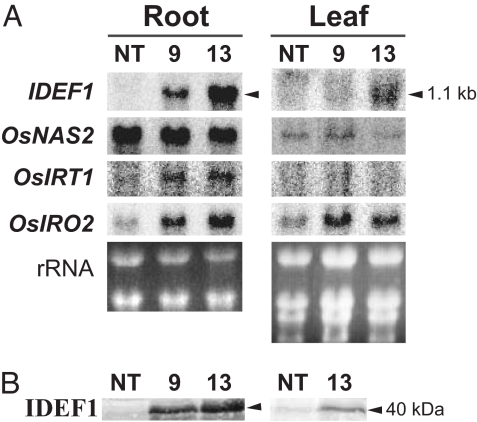
Northern and Western blot analyses of I2p-IDEF1 plants confirmed that IDEF1 expression was induced strongly in iron-deficient roots and moderately in iron-deficient leaves (Fig. 4), reflecting the induction pattern of the IDS2 promoter in rice (17). In I2p-IDEF1 plants, genes involved in MAs-based iron acquisition were induced, including OsNAS2 (28), at levels similar to or lower than in NT plants (Fig. 4A and data not shown), presumably as a combined consequence of the initial transactivation and subsequent regulation reflecting the improved iron-nutrition status caused by IDEF1. The plants showed transactivation of OsIRT1, a ferrous transporter gene (29, 30), and OsIRO2, an iron deficiency-induced bHLH transcription factor gene (10, 11) (Fig. 4A). Thus, enhanced IDEF1 expression is thought to lead to tolerance of iron deficiency through the regulation of iron acquisition-related genes, directly through the binding of IDEF1 to IDE1-like elements and also indirectly through the induction of OsIRO2 (SI Fig. 8). Further studies will shed light on the mechanism of the recognition of specific gene sets that are induced or repressed by IDEF1 and on possible cross-talk among micronutrient homeostasis, hormonal regulation, and seed maturation regulated by the ABI3/VP1 family of transcription factors.
Fig. 4.
Gene expression profiles in rice IDEF1 transformants. (A) Northern blot analysis of IDEF1 and iron deficiency-induced genes [OsNAS2 (28), OsIRT1 (29, 30), and OsIRO2 (10, 11)] in roots and leaves of I2p-IDEF1 and NT plants at 7 days of iron deficiency in hydroponic culture. Ten micrograms of total RNA were loaded per lane. IDEF1-hybridizing bands of about the size of the estimated full-length IDEF1 transgene (lacking untranslated regions; 1.1 kb) are indicated. (B) Western blot analysis of IDEF1 in roots and leaves of I2p-IDEF1 and NT plants on day 7 with 40 μg of total protein loaded per lane. Bands about the size of the predicted full-length IDEF1 (40 kDa) are indicated.
IDEF1 as a Key Component in the Regulation of Iron-Deficiency Responses and Tolerance.
In contrast to previously reported transcription factors related to plant nutrition, IDEF1 binds specifically to the functionally identified cis-acting element IDE1, which is thought to be functional in a wide range of plant species (8, 16–18). Transcripts of IDEF1 are constitutively expressed, and its levels do not increase under iron deficiency (Fig. 1F). Thus, IDEF1 may constitute a key component that transmits the iron-deficiency signal at an early stage. Interestingly, IDEF1 belongs to an uncharacterized subfamily of the plant-specific transcription-factor family ABI3/VP1 (Fig. 1B). Furthermore, IDEF1 binds effectively to the CATGC sequence (Fig. 1; SI Fig. 5), enabling it to recognize IDE1, which lacks the canonical Sph/RY minimal sequence (CATGCA) required for efficient binding with other characterized ABI3/VP1 transcription factors (20, 21). These findings suggest the acquisition of properties by plants in response to various environmental stimuli by modulating the backbone structure of plant-specific transcription factors and cis-acting sequences.
Graminaceous crops constitute a major part of the world's food supply, and rice and maize are particularly susceptible to iron deficiency. Previously, we produced rice plants with enhanced tolerance to low iron availability due to the introduction of barley HvNAAT genes (31) or an engineered ferric-chelate reductase gene, refre1–372 (32). However, the genetic enhancement of a wide range of related genes requires the manipulation of basal regulatory systems, including transcription factors. Supporting this notion, I2p-IDEF1 rice plants exhibited enhanced tolerance to low iron availability (Fig. 3). Recently, we revealed that the overexpression of OsIRO2 also confers tolerance to iron deficiency (ref. 11; unpublished results). Further manipulation of IDEF1, in combination with IRO2 and other as-yet-unknown factors, may provide plant lines that carry favorable traits for use in problem soils.
Materials and Methods
The primers and oligomers used for subcloning, EMSA and Northern blot analysis are shown in SI Table 1.
Yeast LacZ Assay.
To detect IDE1-binding activity in vivo, a yeast LacZ assay was performed by using the Matchmaker One-Hybrid system (Clontech) according to the Yeast Protocols Handbook (Clontech). A double-stranded DNA fragment containing three tandem copies of IDE1 (IDE1 × 3) and two tandem copies of IDE2-IDE1 (IDE2-IDE1 × 2) were synthesized by annealing sense oligomers and antisense primers followed by in-filling using DNA polymerase. These fragments were inserted into the pLacZi vector (Clontech), which was transformed into yeast (Saccharomyces cerevisiae strain YM4271; Clontech). To express ABI3/VP1-family genes in yeast, full-length cDNA clones were acquired from the Rice Full-Length cDNA Database (KOME), and the ORFs of these cDNAs were amplified by PCR. The fragments were inserted into pGAD424 (Clontech), constructing the ABI3/VP1-family genes in-frame downstream of GAL4 activation domain. These fusion plasmids, as well as pGAD424 itself (pGAD), were used in a quantitative LacZ assay.
EMSA.
Whole or partial ORF sequences of the ABI3/VP1 family, corresponding to amino acids 501–726 of OsVP1, 141–362 of IDEF1, 151–433 of AK072874, 1–289 of AK109920, and 287–672 of AK101356, all containing the B3 DNA-binding region, were amplified by PCR and inserted into pMAL-c2 (New England Biolabs), constructing the transcription factor gene fragments in-frame downstream of the MBP gene. These fusion plasmids, as well as pMAL-c2 itself (which expresses a MBP-LacZ fusion), were introduced into the Escherichia coli strain XL1-Blue. Proteins used for EMSA were obtained by using the MBP fusion system according to the manufacturer's instructions (New England Biolabs). EMSA was carried out as described (10) except for a slight modification in the binding buffer composition. One hundred nanograms of MBP-fused IDEF1 or VP1 homologs was incubated with 15 mM Hepes (pH 7.5), 51 mM KCl, 6% glycerol, 0.05% Igepal CA-630 (Sigma), 200 ng of poly (dI-dC)2 (Amersham), and 0.5–1.0 ng of the IDE1 probe (SI Table 1). For competition analysis, both of the IDE1 sequences in the IDE1 probe were substituted with the derivatives shown in Fig. 1A and SI Fig. 5A.
Northern Blot Analysis.
For the detection of endogenous IDEF1 transcripts, rice (cv. Nipponbare) seedlings were grown hydroponically (8, 10). Transgenic tobacco and rice plants were grown as described below. Total RNA was extracted from roots or leaves using the SDS–phenol method and subjected to 32P-based Northern blot analysis as described (7), except that the final washing at 65°C was omitted. The full-length ORF of IDEF1 was used as a probe. For the OsNAS2 probe, a gene-specific region (28) was amplified by PCR. For the OsIRT1 probe, an OsIRT1 fragment that hybridizes with OsIRT1 and OsIRT2 was amplified by PCR (SI Table 1). For the OsIRO2 probe, a 1.1-kb region of an OsIRO2 EST (10) was used.
Subcellular Localization of IDEF1.
A linker sequence containing a BglII site was introduced between the SalI and NcoI sites of CaMV35S-sGFP(S65T)-NOS3′ (kindly provided by Y. Niwa, University of Shizuoka, Shizuoka, Japan). The ORF of IDEF1 was amplified by PCR and inserted into the above GFP vector to construct an in-frame fusion of IDEF1-GFP. Transient gene expression in onion (Allium cepa) epidermal cells and fluorescence observation were carried out as described (33).
Generation, Growth Conditions, and GUS Assay of Transgenic Tobacco.
The IDE1-IDE1 fragment used for tobacco transformation consisted of two IDE1 sequences flanked on both sides of the −235/−154 region of the IDS2 gene promoter, which was amplified by PCR and inserted into pBI101d46 (16) upstream of the uidA (GUS) gene. To introduce IDEF1 into plants, the IDEF1 ORF (1,089 bp) was amplified by PCR and replaced with the GUS gene of pIG121Hm (34) to produce the 35S-IDEF1 binary vector. For a vector control (VC) for double transformation of tobacco, the 35S-GUS cassette of pIG121Hm was excised by digesting with HindIII and SalI followed by self-ligation. Tobacco (Nicotiana tabacum L. cv. Petit-Havana SR1) leaf discs were subjected to Agrobacterium-mediated transformation to introduce the IDE1-IDE1-GUS construct. After kanamycin screening, T1 leaves of a representative line were used to further introduce 35S-IDEF1 or VC, and double transformants were obtained by screening by using hygromycin B (50 mg· liter−1). The T1 double transformants were grown as described (16) with slight modifications. At 16–17 days after germination on Murashige and Skoog (MS) medium containing hygromycin B, plantlets were transplanted into new MS solid medium containing or lacking iron. Whole roots and the youngest and second-youngest leaves of two plantlets of each line were harvested 14 days after transplanting and used for a fluorometric GUS assay (16), Western blot analysis, and leaf chlorophyll measurement performed by using a SPAD-502 chlorophyll meter (Konica Minolta).
Western Blot Analysis.
Total protein was extracted from roots and leaves of transgenic tobacco and rice plants by using the trichloroacetic acid–acetone method. A rabbit anti-IDEF1 polyclonal antibody was produced (Hokudo) by using an IDEF1 (141–362)-MBP fusion protein after cleavage and elimination of MBP. The IgG fraction of the antiserum (Protein A purification, Hokudo) was used for Western blot analysis with a goat anti-rabbit IgG (H+L) horseradish peroxidase-conjugate secondary antibody (Bio-Rad) and staining with diaminobenzidine.
Generation and Growth Conditions of Transgenic Rice.
The IDS2 promoter-IDEF1 binary vector (I2p-IDEF1) was constructed by inserting the IDEF1 ORF (1,089 bp) isolated above into the construct I2 (17), which possesses a 1,663-bp promoter region of the barley IDS2 gene in the pIG121Hm vector. The 35S-IDEF1 and I2p-IDEF1 constructs were introduced into rice (cv. Tsukinohikari) via Agrobacterium-mediated transformation as described (7). The T1 seeds and nontransformants were germinated and grown for 16 days. After an acclimation period of 3 days, the plantlets were transferred to hydroponic culture (8). After 7 days for NT and I2p-IDEF1 plants, or after 7–14 days for 35S-IDEF1 plants, when the shoot height reached ≈20–30 cm, iron deficiency was induced by transplanting seedlings into culture solution without Fe(III)-EDTA or pH adjustment. The nutrient solution was renewed on days 4 and 7. The SPAD meter index of the largest or the youngest leaf was measured. Roots and shoots were harvested on days 7 and 12 and used for Northern and Western analyses and iron concentration measurement. For the growth assay on a calcareous soil, 10 seeds of the T1 transformant or a NT were germinated in a pot filled with 500 g of calcareous soil [pH 8.5 (31, 32)] with 1 g of the controlled-release NPK-type fertilizer CRF-NPK (Chisso Asahi) to supply macronutrients. The pots were watered daily. At day 17, the pots were waterlogged and grown for an additional 8 days, at which point the shoots of the plantlets were used for iron concentration measurements.
Determination of Iron Concentrations.
The harvested plant segments were dried for 2 days at 80°C, and 100- to 200-mg portions were then wet-ashed with 4 ml of 4.4 M HNO3 and 6.5 M H2O2 for 20 min at 230°C using a MarsXpress oven (CEM). Iron concentrations were measured by using inductively coupled plasma atomic emission spectrometry (SPS1200VR; Seiko) at 238.204 nm.
Supplementary Material
Acknowledgments
We thank Dr. Yoshiaki Nagamura of the Rice Genome Project (Tsukuba, Japan) and the National Institute of Agrobiological Sciences DNA Bank (Tsukuba, Japan) for providing the rice cDNA clones and Dr. Yasuo Niwa for providing the CaMV35S-sGFP(S65T)-NOS3′ plasmid. We also thank Drs. Haruhiko Inoue for assistance with plant culture, Pax Blamey and Khurram Bashir for critically reading the manuscript, and Toshihiro Yoshihara for valuable suggestions on the gain-of-function analysis of IDE1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707010104/DC1.
References
- 1.Marschner H. Mineral Nutrition of Higher Plants. 2nd Ed. London, UK: Academic; 1995. [Google Scholar]
- 2.Römheld V, Marschner H. Plant Physiol. 1986;80:175–180. doi: 10.1104/pp.80.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takagi S. Soil Sci Plant Nutr. 1976;22:423–433. [Google Scholar]
- 4.Mori S, Nishizawa N. Plant Cell Physiol. 1987;28:1081–1092. [Google Scholar]
- 5.Shojima S, Nishizawa NK, Fushiya S, Nozoe S, Irifune T, Mori S. Plant Physiol. 1990;93:1497–1503. doi: 10.1104/pp.93.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mori S. Curr Opin Plant Biol. 1999;2:250–253. doi: 10.1016/S1369-5266(99)80043-0. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi T, Nakanishi H, Takahashi M, Kawasaki S, Nishizawa NK, Mori S. Planta. 2001;212:864–871. doi: 10.1007/s004250000453. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi T, Suzuki M, Inoue H, Itai RN, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. J Exp Bot. 2005;56:1305–1316. doi: 10.1093/jxb/eri131. [DOI] [PubMed] [Google Scholar]
- 9.Bashir K, Inoue H, Nagasaka S, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. J Biol Chem. 2006;281:32395–32402. doi: 10.1074/jbc.M604133200. [DOI] [PubMed] [Google Scholar]
- 10.Ogo Y, Itai RN, Nakanishi H, Inoue H, Kobayashi T, Suzuki M, Takahashi M, Mori S, Nishizawa NK. J Exp Bot. 2006;57:2867–2878. doi: 10.1093/jxb/erl054. [DOI] [PubMed] [Google Scholar]
- 11.Ogo Y, Itai RN, Nakanishi H, Kobayashi T, Takahashi M, Mori S, Nishizawa NK. Plant J. 2007;51:366–377. doi: 10.1111/j.1365-313X.2007.03149.x. [DOI] [PubMed] [Google Scholar]
- 12.Ling HQ, Bauer P, Bereczky Z, Keller B, Ganal M. Proc Natl Acad Sci USA. 2002;99:13938–13943. doi: 10.1073/pnas.212448699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colangelo EP, Guerinot ML. Plant Cell. 2004;16:3400–3412. doi: 10.1105/tpc.104.024315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jakoby M, Wang HY, Reidt W, Weisshaar B, Bauer P. FEBS Lett. 2004;577:528–534. doi: 10.1016/j.febslet.2004.10.062. [DOI] [PubMed] [Google Scholar]
- 15.Vorwieger A, Gryczka C, Czihal A, Douchkov D, Tiedemann J, Mock HP, Jakoby M, Weisshaar B, Saalbach I, Baumlein H. Planta. 2007;226:147–158. doi: 10.1007/s00425-006-0476-9. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi T, Nakayama Y, Itai RN, Nakanishi H, Yoshihara T, Mori S, Nishizawa NK. Plant J. 2003;36:780–793. doi: 10.1046/j.1365-313x.2003.01920.x. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi T, Nakayama Y, Takahashi M, Inoue H, Nakanishi H, Yoshihara T, Mori S, Nishizawa NK. Soil Sci Plant Nutr. 2004;50:1167–1175. [Google Scholar]
- 18.Ducos E, Fraysse ÅS, Boutry M. FEBS Lett. 2005;579:6791–6795. doi: 10.1016/j.febslet.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki M, Kao CY, McCarty DR. Plant Cell. 1997;9:799–807. doi: 10.1105/tpc.9.5.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reidt W, Wohlfarth T, Ellerström M, Czihal A, Tewes A, Ezcurra I, Rask L, Bäumlein H. Plant J. 2000;21:401–408. doi: 10.1046/j.1365-313x.2000.00686.x. [DOI] [PubMed] [Google Scholar]
- 21.Mönke G, Altschmied L, Tewes A, Reidt W, Mock HP, Bäumlein H, Conrad U. Planta. 2004;219:158–166. doi: 10.1007/s00425-004-1206-9. [DOI] [PubMed] [Google Scholar]
- 22.Hattori T, Terada T, Hamasuna ST. Plant Mol Biol. 1994;24:805–810. doi: 10.1007/BF00029862. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki M, Wang HHY, McCarty DR. Plant Physiol. 2007;143:902–911. doi: 10.1104/pp.106.092320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsukagoshi H, Morikami A, Nakamura K. Proc Natl Acad Sci USA. 2007;104:2543–2547. doi: 10.1073/pnas.0607940104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higuchi K, Tani M, Nakanishi H, Yoshihara T, Goto F, Nishizawa NK, Mori S. Biosci Biotechnol Biochem. 2001;65:1692–1696. doi: 10.1271/bbb.65.1692. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi T, Yoshihara T, Itai RN, Nakanishi H, Takahashi M, Mori S, Nishizawa NK. Plant Physiol Biochem. 2007;45:262–269. doi: 10.1016/j.plaphy.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 27.Nozoye T, Inoue H, Takahashi M, Ishimaru Y, Nakanishi H, Mori S, Nishizawa NK. Plant Mol Biol. 2007;64:35–47. doi: 10.1007/s11103-007-9132-4. [DOI] [PubMed] [Google Scholar]
- 28.Inoue H, Higuchi K, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. Plant J. 2003;36:366–381. doi: 10.1046/j.1365-313x.2003.01878.x. [DOI] [PubMed] [Google Scholar]
- 29.Bughio N, Yamaguchi H, Nishizawa NK, Nakanishi H, Mori S. J Exp Bot. 2002;53:1677–1682. doi: 10.1093/jxb/erf004. [DOI] [PubMed] [Google Scholar]
- 30.Ishimaru Y, Suzuki M, Tsukamoto T, Suzuki K, Nakazono M, Kobayashi T, Wada Y, Watanabe S, Matsuhashi S, Takahashi M, et al. Plant J. 2006;45:335–346. doi: 10.1111/j.1365-313X.2005.02624.x. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi M, Nakanishi H, Kawasaki S, Nishizawa NK, Mori S. Nature Biotechnol. 2001;19:466–469. doi: 10.1038/88143. [DOI] [PubMed] [Google Scholar]
- 32.Ishimaru Y, Kim S, Tsukamoto T, Oki H, Kobayashi T, Watanabe S, Matsuhashi S, Takahashi M, Nakanishi H, Mori S, et al. Proc Natl Acad Sci USA. 2007;104:7373–7378. doi: 10.1073/pnas.0610555104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizuno D, Higuchi K, Sakamoto T, Nakanishi H, Mori S, Nishizawa NK. Plant Physiol. 2003;132:1989–1997. doi: 10.1104/pp.102.019869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiei Y, Ohta S, Komari T, Kumashiro T. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.