Abstract
The crystal structure of an HIV-1 trans-activation response region (TAR) RNA fragment containing the binding site for the trans-activation protein Tat has been determined to 1.3-Å resolution. In this crystal structure, the characteristic UCU bulge of TAR adopts a conformation that is stabilized by three divalent calcium ions and differs from those determined previously by solution NMR. One metal ion, crucial to the loop conformation, binds directly to three phosphates in the loop region. The structure emphasizes the influence of metal ion binding on RNA structure and, given the abundance of divalent metal ion in the cell, raises the question of whether metal ions play a role in the conformation of TAR RNA and the interaction of TAR with Tat and cyclin T in vivo.
The trans-activation response region (TAR) of the HIV is a 59-nt RNA stem loop located at the 5′ termini of viral transcripts that plays an important role in regulating HIV gene expression. Upon binding the regulatory protein Tat and the host cellular proteins cyclin T and CDK9, the TAR hairpin stabilizes RNA polymerase II to allow viral transcription (1–6). Only the apical portion of the stem loop from positions +19 to +42 is required for transcriptional activation in vivo (7). Extensive mutagenesis studies have shown that the region of TAR important for binding Tat involves a set of nucleotides that surround a characteristic UCU nucleotide bulge; important residues include U23, the first bulged nucleotide, and the two base pairs that immediately precede and follow the bulge (8–12). Electrophoretic mobility shift experiments have shown that Tat binds to TAR through an arginine-rich domain located from residues 49–57 of the 72-aa protein (9, 11, 13–18) and that a “core” domain spanning residues 32–48 is important in enhancing binding specificity (17). Given the critical roles that these elements have in HIV transcription, inhibition of the interaction between Tat and TAR may offer a drug design target, which would require a detailed knowledge of the protein–RNA recognition elements utilized.
Previous NMR studies have shown that the HIV-1 TAR RNA hairpin can adopt two major conformations. In the absence of ligand, the bulged nucleotides stack within the RNA stem, severely distorting its helical continuity (19, 20). When either argininamide or a Tat peptide containing the basic domain bind to the TAR RNA, the bulge nucleotides loop out of the remaining stem, thereby allowing the upper and lower helical regions to stack coaxially (19, 21). Although the specific interactions between HIV-1 Tat and TAR are still unknown, the structure of the analogous BIV Tat/TAR complex reveals that a 14- or 17-residue peptide encompassing the Tat basic region forms a β-hairpin motif that binds in the TAR major groove (22, 23).
In an effort to examine the RNA structural motifs that govern Tat recognition in atomic detail and to explore the influences of divalent metal ions, we have solved the three-dimensional crystal structure of a 27-nt TAR RNA fragment at 1.3-Å resolution. Interestingly, the characteristic UCU bulge adopts a novel conformation that is stabilized by three bound calcium ions. The metal ion-bound bulge structure is consistent with transient electric birefringence studies, which showed previously that distorted RNA molecules containing U-rich bulges straighten in the presence of Mg2+ (24, 25). Metal ions play important roles in stabilizing a variety of RNA structural motifs by forming both inner-sphere and outer-sphere (through water) ligand interactions with RNA-functional groups. For example, the crystal structures of the Tetrahymena group I intron P4–P6 domain (26, 27) and a 5S rRNA domain (28) show bound metal cations that stabilize bulge and loop regions, respectively. The structure of TAR RNA with bound calcium ions reveals a different conformation of the RNA bulge motif than was observed for the metal ion-free structure in solution.
MATERIALS AND METHODS
RNA Purification and Crystallization.
All RNA oligonucleotides used in this study were synthesized by the Yale Keck Microchemical Facility and were deprotected and purified as described previously (29). To anneal the TAR fragment duplex, equal volumes of 2-mM solutions of each RNA strand were mixed and heated to 65°C for 10 min in annealing buffer (5 mM MgCl2/25 mM sodium cacodylate, pH 6.0) and cooled slowly to room temperature. Crystals were obtained by the hanging-drop method in which 1–2 μl of a 1-mM TAR solution in annealing buffer was mixed with an equal volume of a precipitant buffer containing 20% PEG 4000/200 mM NH4Cl/100 mM CaCl2/50 mM sodium cacodylate, pH 6.0, and equilibrated over the precipitant buffer at 19°C. Crystals typically appear within 1 week and belong to the triclinic space group P1 with unit cell dimensions of a = 27.1 Å, b = 27.1 Å, c = 31.6 Å, α = 69.2°, β = 77.6°, γ = 63.1°. Before data collection, crystals were soaked in a cryosolvent containing precipitant buffer plus 20% ethylene glycol for 20 min and then flash-frozen in liquid propane.
Data Collection and Crystal Structure Determination.
A high-resolution native data set to 1.3-Å resolution was collected at the Cornell High Energy Synchrotron Source (CHESS) A1 beamline by using a 2K charge-coupled device (CCD). Because of a large number of overloaded intensities in the low-resolution data shell, this data set was merged with the intensities from a data set obtained from a separate crystal and collected on an R-AXIS IV imaging plate detector system mounted on a Rigaku RU-300 rotating anode. Because of the high degree of isomorphism between the two crystals, the two data sets merged with good statistics. Intensities were calculated and merged by using the programs denzo and scalepack (30).
The structure was solved by multiple isomorphous replacement using anomalous scattering (MIRAS) with two TAR derivatives (BR1 and BR2) in which 5-bromo-deoxycytosine (5Br-dC) was substituted for selected cytosines. The BR1 derivative contained a 5Br-dC substitution at position C45. BR2 contained 5Br-dC substitutions at both the C19 and C45 positions. A complete anomalous diffraction data set was collected by using the inverse-beam strategy on a BR1 derivative crystal at the bromine absorption maxima (λ = 0.912 Å) at the CHESS F2 beamline equipped with a 2K CCD. The relative positions of the two bromine atoms in the BR2 derivative were deduced from a 7σ peak arising from the bromine cross-vector that was observed in the BR2 native Patterson map. Heavy atom parameters were refined and experimental phases calculated by using mlphare (31); phasing statistics are shown in Table 1.
Table 1.
Data collection, phasing, and refinement statistics
| Native* | BR1 | BR2 | |
|---|---|---|---|
| Resolution, Å | 20–1.3 | 20–2.4 | 20–1.8 |
| Reflections | 17,465 | 4,833 | 6,283 |
| % complete (last shell) | 95.0% (91.8%) | 83.4% (43.6%) | 91.6% (68.2%) |
| Rmerge (last shell) | 6.0% (26.7%) | 2.5%† (2.7%)† | 3.5% (5.7%) |
| Anomalous data | No | Yes | No |
| Phasing statistics | |||
| Number of bromine sites | 1 | 2 | |
| Resolution, Å | 20–2.4 | 20–2.0 | |
| Phasing power | 2.38 | 1.72 | |
| RCullis (iso) | 0.58 | 0.64 | |
| RCullis (ano) | 0.78 | ||
| Combined figure of merit (20 Å–2.0 Å) | 0.52 | ||
| Refinement statistics | |||
| R-factor (15,734 reflections) | 12.6% | ||
| Free R-factor (1,731 reflections) | 17.7% | ||
| Number of atoms | 749 | ||
| Number of water molecules | 178 | ||
| Number of calcium ions | 4 | ||
| rmsd bond lengths | 0.009 Å | ||
| rmsd bond angle lengths | 0.025 Å |
Rmerge is defined as Σ|I − 〈I〉|/ΣI. Phasing power is defined as rms FH/rms closure error. RCullis (iso) = ratio of lack of closure to isomorphous difference. RCullis (ano) = ratio of lack of closure to anomalous difference. Figure of merit is defined as cos〈σ(Δφ)〉. R-factor = Σ∥Fo| − |Fc∥/Σ|Fo|, where |Fo| and |Fc| are the observed and calculated structure factors, respectively. Free R-factor is same as R-factor but calculated with 10% of the reflections excluded from refinement.
Native data set was obtained by merging data from two crytals as described in Materials and Methods.
Number reflects Bijvoet pairs treated separately.
The molecular graphics program o (32) was used to fit atom positions into electron density maps calculated with MIRAS phases. The TAR RNA coordinates were refined against the experimental data by using the simulated annealing algorithms contained in “Crystallography and NMR System” (33) in which bulk solvent and anisotropic B corrections were applied to the observed native amplitude set. When the Rfree of the model had dropped to 23.1%, the structure was subjected to restrained anisotropic temperature factor refinement by using shelx-97 (34), which also allowed for the inclusion of fixed hydrogen atoms in the model.
RESULTS
Overall TAR RNA Structure.
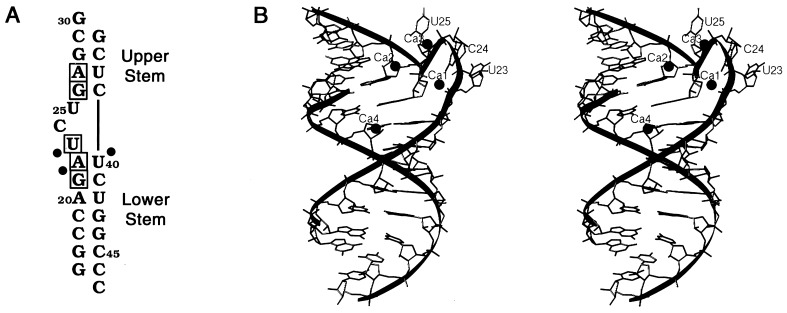
To obtain suitable crystals of the TAR RNA stem, a series of RNA duplex molecules of varying lengths that contained the TAR sequence was constructed and screened for optimal crystal growth. One fragment, shown in Fig. 1A, formed well ordered crystals that allowed high-resolution structure determination by using MIRAS methods. The structure has been refined to a final crystallographic R-factor of 12.6% and a free R-factor of 17.7% (Table 1).
Figure 1.
(A) Sequence of the TAR RNA duplex used in this study. The nucleotides that are important for Tat binding are boxed. Solid circles represent phosphates whose modification disrupts interactions with Tat (17, 35, 36). G16, C30, C46, and C47 are not part of the native TAR sequence. (B) Stereoview of the TAR fragment structure generated with ribbons (37). Spheres depict four calcium ions that bind to the RNA. The nucleotides comprising the bulge are extruded from the stem structure, allowing the upper and lower stem regions to stack linearly.
The TAR RNA fragment in these crystals is a nearly continuous helix formed by the upper and lower stem regions of Watson–Crick base pairs (Fig. 1B). The three bulged nucleotides, U23, C24, and U25, are looped out of the helix, and the U23 and C24 base groups stack on each other. The TAR fragment displays a helical periodicity of 12 bp per turn that is typical of A′-form RNA. The 3′ overhanging G30 nucleotide base pairs with C47 from a symmetry-related molecule allowing the fragments to form uninterrupted helices within the crystal lattice. The presence of the trinucleotide bulge results in a slightly widened major groove in the TAR fragment as compared with that of A-form RNA. The average phosphate–phosphate distance across the major groove in TAR is 12.7 Å whereas the distance in A-RNA is 10.0 Å. Notably, the major groove distortion seen here is not nearly as large as is observed in the RNA structures of the protein-binding domains of the rev response element (38) and 5S rRNA (28).
The majority of the sugar groups in the TAR fragment structure have the C3′-endo conformation characteristic of A-form RNA. Concomitant with the phosphate backbone distortion associated with the UCU bulge, the sugar rings in this region deviate from the standard conformation: U25 is C2′-endo whereas U23 and G26 are both C2′-exo-C3′-endo. Also, the 3′ overhang G30 nucleotide sugar ring adopts a C1′-exo conformation. This peculiar ring pucker may be necessary to allow efficient base pairing interactions between the symmetry-related overhanging 3′ nucleotides.
Metal Ion-Stabilized Bulge Structure.
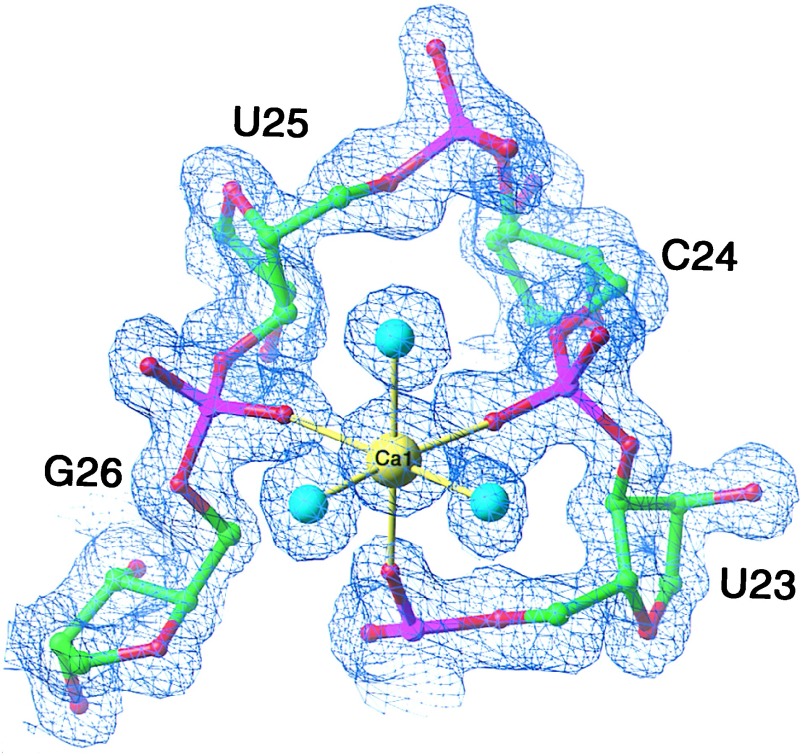
The conformation of the UCU bulge of TAR is stabilized by three divalent metal ions in the crystal structure. Although both Mg2+ and Ca2+ were present in millimolar concentrations in the crystallization buffer, the metal ion-binding sites observed in the structure are best modeled with calcium ions as judged by refinement of their thermal B factors. The 2Fo-Fc electron density map shows one calcium ion (Ca1) to form direct ligands with the pro-Sp phosphate oxygen of U23 and the pro-Rp phosphate oxygens of C24 and G26 (Fig. 2). The octahedrally coordinated Ca1 binds three water molecules directly, two of which interact with the pro-Rp and pro-Sp phosphate oxygens of A22 (Fig. 3).
Figure 2.
The refined model superimposed on a 2Fo-Fc electron density map of the TAR RNA Ca1-binding site contoured at 1.3 σ. Ca1 (yellow sphere) stabilizes the bulge backbone structure through direct ligands with the phosphate oxygens of U23, C24, and G26. Three water molecules, depicted as cyan-colored spheres, complete the octahedral coordination. All Ca2+-phosphate oxygen distances are 2.3 Å. All Ca2+-water distances are 2.4 Å.
Figure 3.
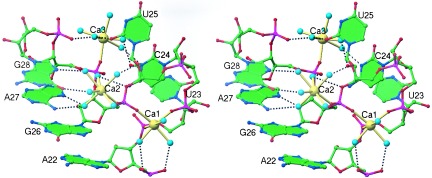
Stereo diagram of the TAR RNA 3-nt bulge region showing the direct and indirect ligands of Ca1, Ca2, and Ca3. Water molecules are colored cyan. Solid yellow bonds denote inner-sphere ligand interactions with the RNA, and black dashed bonds represent outer-sphere coordination. Additional water molecules binding in the bulge region have been omitted for clarity.
In contrast to the Ca1-binding site, calcium ion number 2 (Ca2) and calcium ion number 3 (Ca3) bind to the bulge region through only one direct ligand: Ca2 coordinates the pro-Sp phosphate oxygen of G26, and Ca3 coordinates the pro-Sp phosphate oxygen of A27 (Fig. 3). Ca2 and Ca3 are each hexahydrated, resulting in a coordination number of 7 with the ligands arranged in distorted pentagonal bipyramid configurations. Whereas the RNA ligands of Ca1 involve solely phosphate oxygen atoms, Ca2 and Ca3 form a larger array of outer-sphere ligand interactions with backbone, sugar, and base functional groups of C24, U25, G26, A27, and G28 (Fig. 3). In addition, Ca3 makes two outer-sphere interactions with a symmetry-related molecule (not shown in Fig. 3), which presumably stabilize the crystal lattice.
Based on the current structural data alone, we cannot differentiate between metal ion-binding sites that are important for stabilizing the bulge conformation and those that are necessary to maintain the crystal lattice. Furthermore, we cannot determine the relative affinity of each calcium ion to TAR RNA, although we can report that all metal ion sites remain fully substituted after soaking TAR RNA crystals in a solution containing 10 mM CaCl2 (data not shown). Interestingly, hydrodynamic measurements indicate that Mg2+ straightens RNA molecules that contain U3 trinucleotide bulges, but not those that have A3 bulges (24). These results suggest that metal ion stabilization is sequence-specific and requires at least one divalent metal ion to interact with the pyrimidine base groups, as Ca3 does in this structure, to induce the looped-out bulge conformation.
Additionally, the slight distortion associated with the bulge allows a fourth calcium ion (Ca4) to bind in the major groove of the TAR fragment (Fig. 1B). This metal ion is octahydrated and makes no direct coordination with the RNA, but does engage in outer-sphere ligand interactions with the O6 and N7 atoms of G21, the N6 atom of A22, and the O6 atom of G26. Both hydrated magnesium ion and cobalt (III) hexamine have been observed to bind to G residues involved in noncanonical base pairs in distorted major grooves of RNA (26, 27, 39, 40).
Furthermore, the calcium ion-bound bulge in TAR RNA provides a second example of a metal ion-stabilized bulge region in RNA. Similarly, the structure of the P4–P6 domain of the group I intron contains an A-rich 5-nt bulge, which adopts a more stable structure upon binding divalent metal ions than its metal ion-free conformation in solution (26, 27, 41). In this A-rich bulge motif, two Mg2+ ions each directly ligand three different phosphate oxygens to stabilize a “corkscrew” turn in the group I intron P5a helix.
DISCUSSION
Comparison with Other TAR RNA Structures.
Previous structural studies have shown that the apical portion of HIV-1 TAR RNA stem loop adopts different conformations in the presence or absence of a guanidinium-containing ligand. Using NMR spectroscopic methods, Williamson and colleagues (19) found that in the absence of any ligand, U23 stacks on A22 resulting in a bent helical structure for the TAR stem loop. However, in the presence of 6 mM argininamide, the bulged nucleotides loop out from the remaining helix, which allows the upper and lower stems to stack coaxially. In the resulting model of the “bound” TAR, U23 is proposed to form a base triple interaction with A27 and U38 even though no direct NOE measurements between U23 and A27 bases are observed. However, ligand-induced U23⋅A27⋅U38 base triple formation is supported by TAR mutagenesis studies, which show that the U23C mutant does not bind arginine unless the compensatory A27G and U38C mutations are also present, presumably to allow a C23⋅G27⋅C38 interaction (42, 43). Spectroscopic results also reveal that a single guanidinium group of argininamide interacts with the O6 and N7 atoms of G26 and the phosphate groups of A22 (P22) and U23 (P23) (19, 42). Further support for base triple formation upon ligand binding is shown in the solution structure of the analogous HIV-2 TAR, which contains a UU dinucleotide bulge, in complex with argininamide where U23 is found to be within hydrogen-bonding distance of A27 (44).
In another study, Varani and coworkers (21) report an alternate structure of TAR RNA in the bound conformation derived from a larger set of NMR experimental constraints. As in the model calculated by Puglisi et al. (19), the U23 base is located adjacent to the major groove nucleotides. However, U23 is too distant from A27 to allow a base triple interaction. The structure of the UCU bulge observed in this study prevents P22, P23, and the G26 base from interacting with solely one arginine residue, as proposed previously (19, 42), because of spatial constraints.
Of the two major conformations observed in the NMR studies, the overall structure of crystalline TAR–calcium complex is more similar to the “bound” conformer in that the bulged nucleotides in the crystal structure are extruded out of the stem region. Likewise, both the TAR crystal structure and the “bound” conformer display similar helical continuity and narrow major groove dimensions. Importantly, the still relatively narrow major groove observed in these structures would not necessarily occlude Tat from binding within the TAR major groove. Based on the NMR structure of the TAR bound form, Aboul-ela et al. (21) postulate that Tat gains access to the major groove of TAR as a consequence of phosphate backbone discontinuity imposed by the bulged nucleotides.
Because the solution structures of TAR have been determined in the absence of divalent metals, it may not be surprising that the conformation of the trinucleotide bulge observed in the crystal structure differs significantly from any of the conformations observed in NMR studies. Fig. 4 compares the bulge conformations observed in the TAR RNA crystal structure with those of the bound form of TAR as determined by Puglisi et al. (19) and Aboul-ela et al. (21) by using NMR methods. The largest difference among the structures appears to be the position of U23, a residue whose mutation reduces Tat affinity both in vitro and in vivo (17, 42). As a consequence of the calcium ion-stabilized phosphate backbone conformation, U23 orients away from the major groove in the TAR RNA crystal structure. Also, the N7 of A27, the potential hydrogen-bonding partner of U23, interacts with Ca2. The crystal structure shows that G26 is still available to engage in a potential bidentate arginine interaction (45) with the Tat basic region (Fig. 3). In all, the comparison between the metal ion-free structure in solution and the metal ion-bound form of crystalline TAR elucidates the significant effect that metal ions have on the structures of small RNA motifs.
Figure 4.
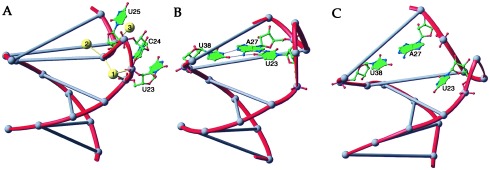
Comparison of the TAR RNA bulge conformations observed in x-ray and NMR studies. The nucleotide backbones of the lower-stem regions of each structure have been superimposed and then translated. For clarity, the Watson–Crick base pairs of the TAR stem are denoted as lines between corresponding phosphate atoms, shown as gray spheres. (A) TAR RNA x-ray structure. Bulged nucleotides U23, C24, and U25 are extruded from the helical stem region. Ca1, Ca2, and Ca3, shown as yellow spheres, stabilize the bulge structure that prevents U23 from interacting with the major groove. (B) The structure of the TAR “bound” conformation as determined by Puglisi et al. (19) by using NMR methods. Although U23, C24, and U25 also are looped out in this structure, U23 forms a base triple interaction with A27⋅U38. Because the C24 and U25 positions are not determined accurately, they hence are not represented. (C) TAR RNA “bound” structure as determined by Aboul-ela et al. (21) also by NMR. Although U23 is positioned adjacent to the major groove, it is too distant to interact with A27.
Biological Implications of the TAR Crystal Structure.
Consistent with previous hydrodynamic studies (24, 25), our structural results show that the TAR RNA bulge region adopts a different conformation upon binding divalent metal ions. Given the abundance of intracellular divalent metal ions, the calcium ion-binding sites observed in the TAR RNA crystal structure raise the possibility that metal ions may play a role in Tat recognition of TAR in vivo. Surprisingly, very little data exist that correlate the effect of divalent metal ion concentration upon the interaction between Tat and TAR, because the in vitro binding and modification studies were done largely in the absence of metal ions. Perhaps more significantly, little is known concerning the influence of divalent metal ions on the binding of full-length Tat and the host protein, cyclin T, to TAR RNA.
In a recent investigation, Zacharias and Hagerman (25) studied the effects of Mg2+-ion concentration upon the bulge-induced bend and arginine affinity of TAR RNA by using transient electric birefringence. Mg2+ (2 mM) reduces the 50° bend angle of apo-TAR RNA by approximately 50%. Although the metal-induced transition is not as large as produced by arginine, where a nearly complete straightening of TAR is observed, the authors speculate that higher Mg2+ concentrations would have induced further straightening. Furthermore, they find that as much as 0.90 mM Mg2+ has no measurable effect on the concentration of arginine at which the midpoint of the structural transition occurs, suggesting that arginine-binding affinity is unaltered by magnesium ion. These results imply that TAR–arginine interactions are not necessarily inhibited in the presence of positively charged metal ions.
Although exact binding constants of Mg2+ to TAR have not been determined in this or any other study, the structural transitions observed in the hydrodynamic measurements that are induced by 2 mM MgCl2 do suggest that TAR can bind Mg2+ at physiological concentrations of this divalent metal ion. Consequently, this implies that in the cell, Tat may initially recognize a metal ion-bound TAR RNA species as well as the metal ion-free structure observed in solution (19, 20).
Unfortunately, the set of conditions required to grow the crystals of TAR, whose structure is reported here, currently limits our ability to correlate the calcium ion-bound structure of TAR with its function in vivo. Optimal crystallization of the TAR stem fragment requires 100 mM CaCl2. Although TAR crystals are stable in only 10 mM CaCl2 and maintain the same bulge structure, this Ca2+ concentration still exceeds the intracellular calcium concentration by several orders of magnitude. However, we note that Mg2+ may substitute for some of the calcium ion sites in vivo and maintain the same structure we observe in our study. On the other hand, the position of U23 in the calcium ion-bound structure is inconsistent with the current biochemical model of the Tat/TAR interaction. Both mutagenesis and chemical modification (15, 17, 35, 46) studies have shown that a U at position 23 is important for the specific binding of both arginine alone and Tat-derived peptides. However, in the TAR crystal structure, the metal-bound backbone conformation of the UCU bulge places U23 in a position where it is not likely to make any direct, base-specific interactions with the basic Tat peptides or form part of an arginine-binding pocket, as has been proposed previously (19, 42), without significant structural rearrangement of the base.
The crystal structure of the HIV-1 TAR RNA stem demonstrates that divalent metal ions can drastically affect the conformation of RNA bulge motifs. Although we cannot directly correlate our results with the function of TAR at this time, the metal ion-bound trinucleotide bulge we observe in the TAR crystal structure may represent a stabilizing motif that will appear in other RNA structures.
Acknowledgments
We thank S. Bellon, G. Cheetham, C. Correll, P. Klosterman, and Y. Shamoo for providing general assistance with the structure determination and Professors D. Crothers, P. Moore, and J. Williamson for critical reading of the manuscript. This work is supported by National Institutes of Health Grant GM39546 to T.A.S. and a postdoctoral fellowship (GM17645) to J.A.I.
ABBREVIATION
- TAR
trans-activation response region
Footnotes
Data deposition: The atomic coordinates described in this paper have been deposited into the Nucleic Acid Data Base, Rutgers, The State University of New Jersey, Piscataway, NJ 08854 (accession code URX075).
References
- 1.Muesing M A, Smith D H, Capon D J. Cell. 1987;48:691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- 2.Feng S, Holland E C. Nature (London) 1988;334:165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- 3.Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M B, Price D H. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mancebo H S Y, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, Flores O. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang X, Gold M O, Tang D N, Lewis D E, Aguilar-Cordova E, Rice A P, Herrmann C H. Proc Natl Acad Sci USA. 1997;94:12331–12336. doi: 10.1073/pnas.94.23.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei P, Garber M E, Fang S-M, Fischer W H, Jones K A. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 7.Jakobovits A, Smith D H, Jakobovits E B, Capon D J. Mol Cell Biol. 1988;8:2555–2561. doi: 10.1128/mcb.8.6.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dingwall C, Ernberg I, Gait M J, Green S M, Heaphy S, Karn J, Lowe A D, Singh M, Skinner M A, Valerio R. Proc Natl Acad Sci USA. 1989;86:6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weeks K M, Ampe C, Schultz S C, Steitz T A, Crothers D M. Science. 1990;249:1281–1285. doi: 10.1126/science.2205002. [DOI] [PubMed] [Google Scholar]
- 10.Roy S, Delling U, Chen C-H, Rosen C A, Sonenberg N. Genes Dev. 1990;4:1365–1373. doi: 10.1101/gad.4.8.1365. [DOI] [PubMed] [Google Scholar]
- 11.Cordingley M G, LaFemina R L, Callahan P L, Condra J H, Sardana V V, Graham D J, Nguyen T M, LeGrow K, Gotlib L, Schlabach A J, Colonno R J. Proc Natl Acad Sci USA. 1990;87:8985–8989. doi: 10.1073/pnas.87.22.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delling U, Reid L S, Barnett R W, Ma M Y-X, Climie S, Sumner-Smith M, Sonenberg N. J Virol. 1992;66:3018–3025. doi: 10.1128/jvi.66.5.3018-3025.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dingwall C, Ernberg I, Gait M J, Green S M, Heaphy S, Karn J, Lowe A D, Singh M, Skinner M A. EMBO J. 1990;9:4145–4153. doi: 10.1002/j.1460-2075.1990.tb07637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calnan B J, Biancalana S, Hudson D, Frankel A D. Genes Dev. 1991;5:201–210. doi: 10.1101/gad.5.2.201. [DOI] [PubMed] [Google Scholar]
- 15.Weeks K M, Crothers D M. Cell. 1991;66:577–588. doi: 10.1016/0092-8674(81)90020-9. [DOI] [PubMed] [Google Scholar]
- 16.Weeks K M, Crothers D M. Biochemistry. 1992;31:10281–10287. doi: 10.1021/bi00157a015. [DOI] [PubMed] [Google Scholar]
- 17.Churcher M J, Lamont C, Hamy F, Dingwall C, Green S M, Lowe A D, Butler P J G, Gait M J, Karn J. J Mol Biol. 1993;230:90–110. doi: 10.1006/jmbi.1993.1128. [DOI] [PubMed] [Google Scholar]
- 18.Long K S, Crothers D M. Biochemistry. 1995;34:8885–8895. doi: 10.1021/bi00027a041. [DOI] [PubMed] [Google Scholar]
- 19.Puglisi J D, Tan R, Calnan B J, Frankel A D, Williamson J R. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 20.Aboul-ela F, Karn J, Varani G. Nucleic Acids Res. 1996;24:3974–3981. doi: 10.1093/nar/24.20.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aboul-ela F, Karn J, Varani G. J Mol Biol. 1995;253:313–332. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 22.Puglisi J D, Chen L, Blanchard S, Frankel A D. Science. 1995;270:1200–1203. doi: 10.1126/science.270.5239.1200. [DOI] [PubMed] [Google Scholar]
- 23.Ye X, Kumar R A, Patel D J. Chem Biol. 1995;2:827–840. doi: 10.1016/1074-5521(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 24.Zacharias M, Hagerman P J. J Mol Biol. 1995;247:486–500. doi: 10.1006/jmbi.1995.0155. [DOI] [PubMed] [Google Scholar]
- 25.Zacharias M, Hagerman P J. Proc Natl Acad Sci USA. 1995;92:6052–6056. doi: 10.1073/pnas.92.13.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cate J H, Gooding A R, Podell E, Zhou K, Golden B L, Kundrot C E, Cech T R, Doudna J A. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 27.Cate J H, Hanna R L, Doudna J A. Nat Struct Biol. 1997;4:553–558. doi: 10.1038/nsb0797-553. [DOI] [PubMed] [Google Scholar]
- 28.Correll C C, Freeborn B, Moore P B, Steitz T A. Cell. 1997;91:705–712. doi: 10.1016/s0092-8674(00)80457-2. [DOI] [PubMed] [Google Scholar]
- 29.Correll C C, Freeborn B, Moore P B, Steitz T A. J Biomol Struct Dyn. 1997;15:165–172. doi: 10.1080/07391102.1997.10508183. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Collaborative Computational Project, No. 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 32.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 33.Brünger, A. T., Adams, P. D., Clore, G. M., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., et al. (1998) Acta Cryst. D, in press. [DOI] [PubMed]
- 34.Sheldrick G M. shelx-97, program for crystal structure refinement. Germany: Univ. of Gottingen; 1997. [Google Scholar]
- 35.Tao J, Frankel A D. Proc Natl Acad Sci USA. 1992;89:2723–2726. doi: 10.1073/pnas.89.7.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pritchard C E, Grasby J A, Hamy F, Zacharek A M, Singh M, Karn J, Gait M J. Nucleic Acids Res. 1994;22:2592–2600. doi: 10.1093/nar/22.13.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carson M. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- 38.Battiste J L, Mao H, Rao N S, Tan R, Muhandiram D R, Kay L E, Frankel A D, Williamson J R. Science. 1996;273:1547–1551. doi: 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- 39.Jack A, Ladner J E, Rhodes D, Brown R S, Klug A. J Mol Biol. 1977;111:315–328. doi: 10.1016/s0022-2836(77)80054-5. [DOI] [PubMed] [Google Scholar]
- 40.Kieft J S, Tinoco I., Jr Structure. 1997;5:713–721. doi: 10.1016/s0969-2126(97)00225-6. [DOI] [PubMed] [Google Scholar]
- 41.Luebke K J, Landry S M, Tinoco I., Jr Biochemistry. 1997;36:10246–10255. doi: 10.1021/bi9701540. [DOI] [PubMed] [Google Scholar]
- 42.Puglisi J D, Chen L, Frankel A D, Williamson J R. Proc Natl Acad Sci USA. 1993;90:3680–3684. doi: 10.1073/pnas.90.8.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tao J, Chen L, Frankel A D. Biochemistry. 1997;36:3491–3495. doi: 10.1021/bi962259t. [DOI] [PubMed] [Google Scholar]
- 44.Brodsky A S, Williamson J R. J Mol Biol. 1997;267:624–639. doi: 10.1006/jmbi.1996.0879. [DOI] [PubMed] [Google Scholar]
- 45.Calnan B J, Tidor B, Biancalana S, Hudson D, Frankel A D. Science. 1991;252:1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- 46.Sumner-Smith M, Roy S, Barnett R, Reid L S, Kuperman R, Delling U, Sonenberg N. J Virol. 1991;65:5196–5202. doi: 10.1128/jvi.65.10.5196-5202.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]