Abstract
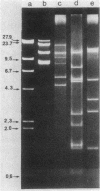
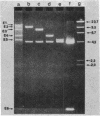
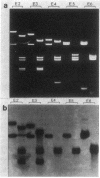
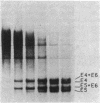
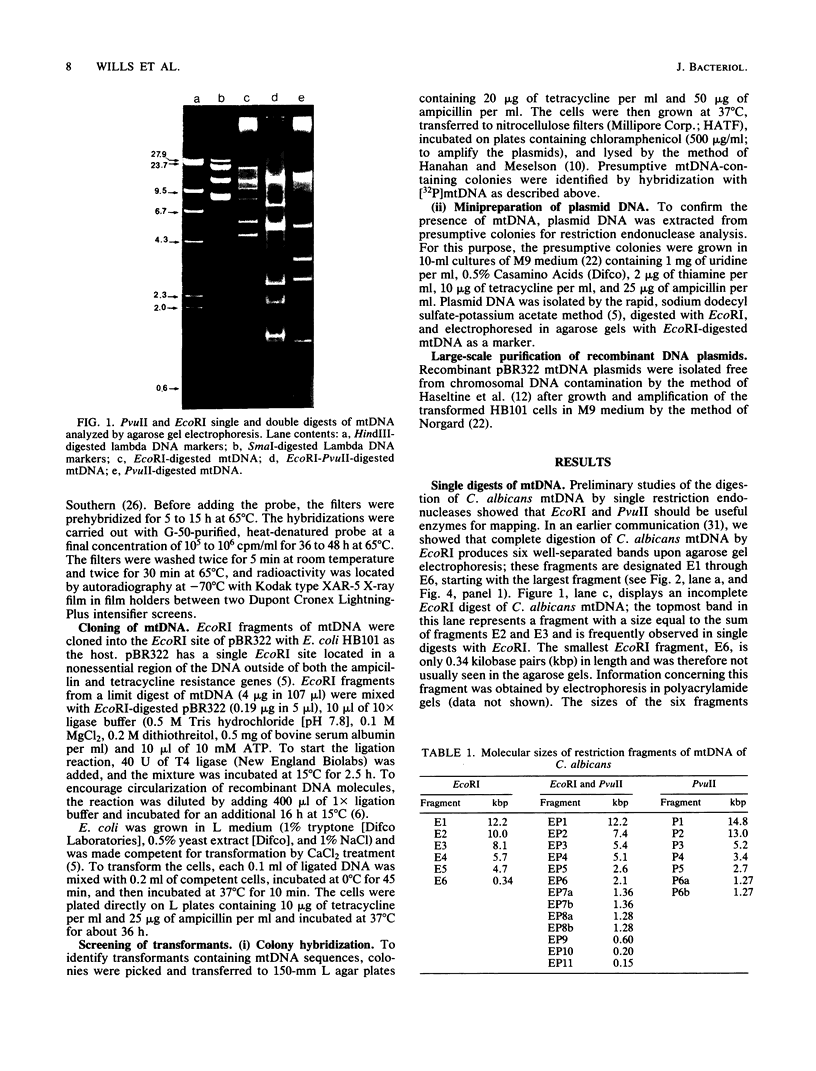
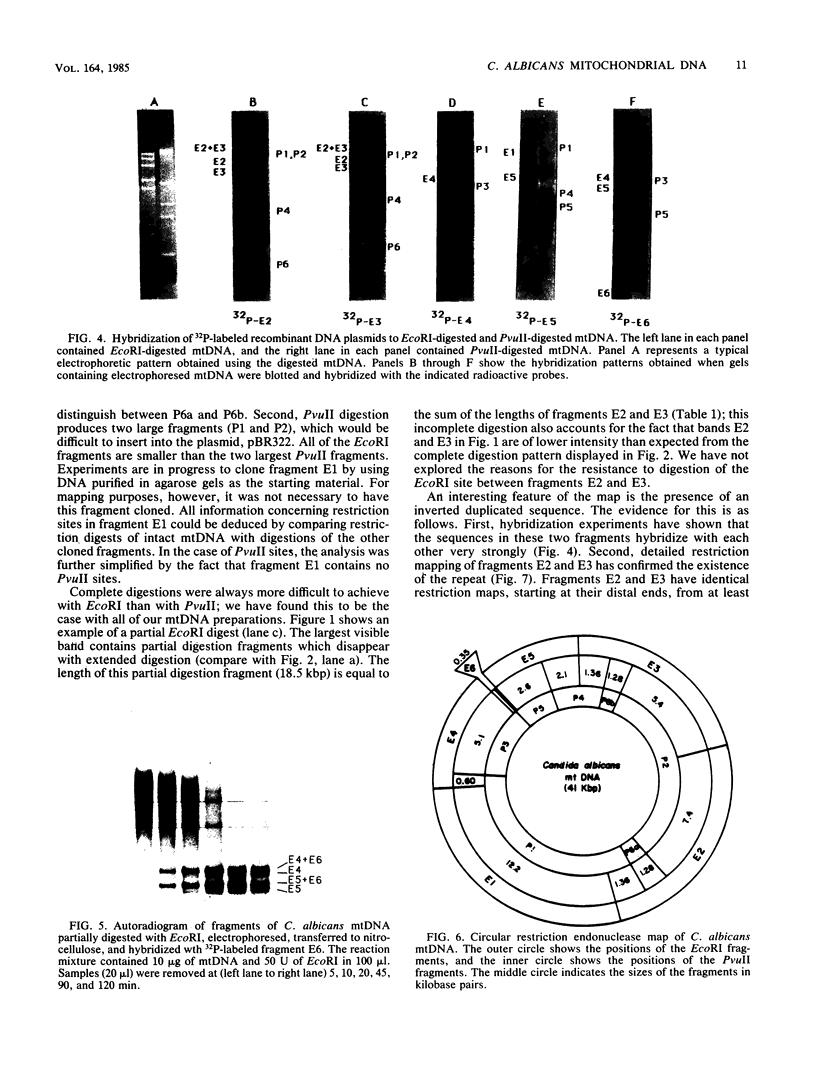
The mitochondrial DNA (mtDNA) of the dimorphic fungus Candida albicans has a molecular size of 41 kilobase pairs as judged by summation of the fragment sizes produced by digestion with restriction endonucleases EcoRI, PvuII, and a combination of both enzymes. Five of the six EcoRI fragments comprising the mitochondrial genome have been cloned into the plasmid vector, pBR322. Restriction mapping revealed a circular map as predicted by previous observations with the electron microscope. The use of nick-translated, purified mtDNA to probe digests of mtDNA from other strains of C. albicans revealed a common restriction pattern. Use of nick-translated, cloned EcoRI fragments to probe digests of mtDNA revealed a large (at least 5 kilobase pairs), inverted duplication as well as a smaller (less than 0.4 kilobase pairs) region of related sequences.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown W. M., George M., Jr, Wilson A. C. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Walker G. D., McArthur C. R., Sriprakash K. S. Partial duplication of the large ribosomal RNA sequence in an inverted repeat in circular mitochondrial DNA from Kloeckera africana. Implications for mechanisms of the petite mutation. J Mol Biol. 1981 Apr 15;147(3):399–415. doi: 10.1016/0022-2836(81)90492-7. [DOI] [PubMed] [Google Scholar]
- Clark-Walker G. D., Sriprakash K. S. Sequence rearrangements between mitochondrial DNAs of Torulopsis glabrata and Kloeckera africana identified by hybridization with six polypeptide encoding regions from Saccharomyces cerevisiae mitochondrial DNA. J Mol Biol. 1981 Sep 25;151(3):367–387. doi: 10.1016/0022-2836(81)90002-4. [DOI] [PubMed] [Google Scholar]
- Dugaiczyk A., Boyer H. W., Goodman H. M. Ligation of EcoRI endonuclease-generated DNA fragments into linear and circular structures. J Mol Biol. 1975 Jul 25;96(1):171–184. doi: 10.1016/0022-2836(75)90189-8. [DOI] [PubMed] [Google Scholar]
- Ferris S. D., Wilson A. C., Brown W. M. Evolutionary tree for apes and humans based on cleavage maps of mitochondrial DNA. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2432–2436. doi: 10.1073/pnas.78.4.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbach R. W., Borst P., Bollen-de Boer J. E., van Bruggen E. F. The organization of ribosomal RNA genes in the mitochondrial DNA of Tetrahymena pyriformis strain ST. Biochim Biophys Acta. 1978 Nov 21;521(1):169–186. doi: 10.1016/0005-2787(78)90260-5. [DOI] [PubMed] [Google Scholar]
- Groot G. S., Flavell R. A., Sanders J. P. Sequence homology of nuclear and mitochondrial DNAs of different yeasts. Biochim Biophys Acta. 1975 Jan 20;378(2):186–194. doi: 10.1016/0005-2787(75)90106-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D., Meselson M. Plasmid screening at high colony density. Gene. 1980 Jun;10(1):63–67. doi: 10.1016/0378-1119(80)90144-4. [DOI] [PubMed] [Google Scholar]
- Haseltine W. A., Lindan C. P., D'Andrea A. D., Johnsrud L. The use of DNA fragments of defined sequence for the study of DNA damage and repair. Methods Enzymol. 1980;65(1):235–248. doi: 10.1016/s0076-6879(80)65033-2. [DOI] [PubMed] [Google Scholar]
- Hudspeth M. E., Shumard D. S., Bradford C. J., Grossman L. I. Organization of Achlya mtDNA: a population with two orientations and a large inverted repeat containing the rRNA genes. Proc Natl Acad Sci U S A. 1983 Jan;80(1):142–146. doi: 10.1073/pnas.80.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet-Vierny C., Begel O., Belcour L. Senescence in Podospora anserina: amplification of a mitochondrial DNA sequence. Cell. 1980 Aug;21(1):189–194. doi: 10.1016/0092-8674(80)90126-9. [DOI] [PubMed] [Google Scholar]
- Johnson M. J., Wallace D. C., Ferris S. D., Rattazzi M. C., Cavalli-Sforza L. L. Radiation of human mitochondria DNA types analyzed by restriction endonuclease cleavage patterns. J Mol Evol. 1983;19(3-4):255–271. doi: 10.1007/BF02099973. [DOI] [PubMed] [Google Scholar]
- Kozłowski M., Stepień P. P. Restriction enzyme analysis of mitochondrial DNA of members of the genus Aspergillus as an aid in taxonomy. J Gen Microbiol. 1982 Mar;128(3):471–476. doi: 10.1099/00221287-128-3-471. [DOI] [PubMed] [Google Scholar]
- Linnane A. W., Lukins H. B. Isolation of mitochondria and techniques for studying mitochondrial biogenesis in yeasts. Methods Cell Biol. 1975;12:285–309. doi: 10.1016/s0091-679x(08)60961-9. [DOI] [PubMed] [Google Scholar]
- Macino G., Scazzocchio C., Waring R. B., Berks M. M., Davies R. W. Conservation and rearrangement of mitochondrial structural gene sequences. Nature. 1980 Nov 27;288(5789):404–406. doi: 10.1038/288404a0. [DOI] [PubMed] [Google Scholar]
- Mannella C. A., Goewert R. R., Lambowitz A. M. Characterization of variant Neurospora crassa mitochondrial DNAs which contain tandem reiterations. Cell. 1979 Dec;18(4):1197–1207. doi: 10.1016/0092-8674(79)90232-0. [DOI] [PubMed] [Google Scholar]
- Norgard M. V. Rapid and simple removal of contaminating RNA from plasmid DNA without the use of RNase. Anal Biochem. 1981 May 1;113(1):34–42. doi: 10.1016/0003-2697(81)90040-3. [DOI] [PubMed] [Google Scholar]
- Riggsby W. S., Torres-Bauza L. J., Wills J. W., Townes T. M. DNA content, kinetic complexity, and the ploidy question in Candida albicans. Mol Cell Biol. 1982 Jul;2(7):853–862. doi: 10.1128/mcb.2.7.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. Gel electrophoresis of restriction fragments. Methods Enzymol. 1979;68:152–176. doi: 10.1016/0076-6879(79)68011-4. [DOI] [PubMed] [Google Scholar]
- Syverson R. E. Variable assimilation of carbon compounds by Candida albicans. J Clin Microbiol. 1981 Jan;13(1):163–166. doi: 10.1128/jcm.13.1.163-166.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Bauzá L. J., Riggsby W. S. Protoplasts from yeast and mycelial forms of Candida albicans. J Gen Microbiol. 1980 Aug;119(2):341–349. doi: 10.1099/00221287-119-2-341. [DOI] [PubMed] [Google Scholar]
- Wallace D. C. Structure and evolution of organelle genomes. Microbiol Rev. 1982 Jun;46(2):208–240. doi: 10.1128/mr.46.2.208-240.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills J. W., Lasker B. A., Sirotkin K., Riggsby W. S. Repetitive DNA of Candida albicans: nuclear and mitochondrial components. J Bacteriol. 1984 Mar;157(3):918–924. doi: 10.1128/jb.157.3.918-924.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R. M., Horrum M. A., Cummings D. J. Are mitochondrial structural genes selectively amplified during senescence in Podospora anserina? Cell. 1982 Jun;29(2):505–515. doi: 10.1016/0092-8674(82)90167-2. [DOI] [PubMed] [Google Scholar]
- Zeman L. J., Lusena C. V. In vivo and in vitro synthesis of yeast mitochondrial DNA. Methods Cell Biol. 1975;12:273–283. doi: 10.1016/s0091-679x(08)60960-7. [DOI] [PubMed] [Google Scholar]