Abstract
We have isolated CaKRE9, a gene from Candida albicans, that is a functional homologue of the Saccharomyces cerevisiae KRE9 gene involved in β-1,6-glucan synthesis. Disruption of the CaKRE9 gene in C. albicans shows that CaKre9p is required for the synthesis or assembly of this fungal polymer. Homozygous null disruptants of CaKRE9 grow poorly on galactose and fail to form hyphae in serum, and, in growth medium containing glucose, the gene is essential. Thus, the CaKRE9 gene product is a potentially useful candidate as a target for fungal-specific drugs.
Opportunistic fungal infections have increased dramatically in the last two decades and have become a significant cause of morbidity and mortality. Yeasts in the genus Candida are the major group causing mycoses in humans ranging from mucosal infections to systemic candidiasis (1–4). Much of the structure, physiology, and metabolism of fungal and animal cells is highly conserved (5). This conservation in cellular function has made it difficult to find agents that selectively discriminate between pathogenic fungi and their human hosts. Moreover, resistance to existing antifungal drugs is becoming a problem of increasing concern (6, 7). Thus, there is an urgent need for specific new targets as a starting point to obtain new antifungal drugs. One focus of attention for such new targets is the fungal extracellular matrix, where the cell wall constitutes an essential, fungal-specific organelle that is absent from human cells. The cell wall of fungi is essential not only in maintaining the osmotic integrity of the fungal cell but also in cell growth, division, and morphogenesis (8, 9). The yeast cell wall is composed of highly mannosylated glycoproteins, chitin, and two homopolymers of glucose, β-1,3-glucan, and β-1,6-glucan (8, 10). The β-1,6-glucan polymer exists as a postranslational modification of cell wall glycoproteins and is essential for fungal cell viability (8–10). It acts as an adhesive, covalently linking glucosylated glycoproteins with the cell wall β-1,3-glucan and chitin polymers in a crosslinked extracellular matrix (11). Cell wall β-1,6-glucan is widespread among fungi occurring in both Ascomycete and Basidiomycete species (12–14). In the dimorphic yeast Candida albicans, β-1,6-glucan is particularly abundant, comprising approximately half of the alkali-insoluble glucan (15, 16) compared with a level of ≈20% found in the yeast Saccharomyces cerevisiae, which has served as the principal organism for studies of this polymer (8–10).
Although the biochemistry of β-1,6-glucosylation is limited, genetic analysis of β-1,6-glucan synthesis has identified many genes required for the process in S. cerevisiae. These genes encode products acting in the endoplasmic reticulum, in the Golgi complex, and at the cell surface (see ref. 10 for a recent review). In the course of this work, the KRE9 gene was identified, and its product was shown to be required for β-1,6-glucan synthesis. KRE9 appears to be a fungal specific gene and encodes a 55-kDa O-glycosylated protein that is found in the extracellular medium when overproduced and is thought to be localized normally to the cell surface, where it participates in glucan synthesis or assembly (17). Disruption of KRE9 in S. cerevisiae leads to serious growth impairment and an altered cell wall containing <20% of the wild-type amount of β-1,6-glucan. The S. cerevisiae genome contains a second gene product, Knh1p, highly similar to Kre9p. Knh1p shares 46% identity with Kre9p and appears to be a functional homolog of Kre9p, as overexpression of KNH1 partially suppressed the severe growth defect of a kre9 null mutant and the level of alkali-insoluble cell wall β-1,6-glucan (18). When overproduced, Knh1p, like Kre9p, can be found in the extracellular culture medium as an O-glycoprotein and is likely also a cell surface protein under conditions of normal expression. The disruption of both the KNH1 and KRE9 genes results in lethality in S. cerevisiae. Transcription of KNH1 is carbon-source and KRE9 dependent. These results suggest that the KRE9 and KNH1 genes are specialized in vivo to function under different environmental conditions (18). The essential nature of the KRE9/KNH1 gene pair and the putative extracellular location of their gene products make these proteins attractive fungal specific targets, a possibility that we have explored further by searching for KRE9 homologue(s) in C. albicans.
MATERIALS AND METHODS
Yeast Strains, Culture Conditions, and Procedures.
The different S. cerevisiae (SEY6210 background) and C. albicans (CAI4 background) strains were grown under standard conditions (yeast extract/peptone/dextrose and yeast nitrogen base/casamino acids). All Ura− C. albicans strains were cultured in presence of uridine (50 μg/ml). Glucose growth studies were performed in yeast nitrogen base medium supplemented with glucose at concentrations of 1%, 0.75%, 0.5%, 0.25%, 0.2%, 0.15%, 0.1%, or 0.05%. The ability of C. albicans strains to grow on different carbon sources was assessed on solid yeast extract/peptone media supplemented with galactose, sucrose, mannose, maltose, fructose, sorbitol, or glycerol at a final concentration of 2%. When cells were scored for growth in the presence of glucose plus another carbon source, the final glucose concentration was 1%. Hyphal growth of different C. albicans strains was induced in presence of galactose and 20% fetal bovine serum at 37°C for up to 18 hr as described (27).
Isolation of the CaKRE9 Gene.
A S. cerevisiae kre9∷HIS3 haploid strain containing a wild-type copy of the S. cerevisiae KRE9 gene (17) on a centromeric LYS2-based pRS317 vector was transformed with a C. albicans genomic library. This library was contained within the multicopy YEp352-plasmid with the URA3 gene as a selectable marker (19). To screen for plasmids that could restore growth to a kre9∷HIS3 mutant, ≈20,000 His3+ Lys2+ Ura3+ cells were replica-plated on minimal medium containing α-aminoadipate as a primary nitrogen source to select for growing cells that had lost the LYS2 plasmid-based copy of KRE9 and that might possess a copy of the complementing CaKRE9 gene. These cells were tested further for loss of the pRS317-KRE9 plasmid by their failure to grow on medium lacking lysine. YEp352-based C. albicans genomic DNA was recovered from cells that required lysine for growth. On retransformation in yeast, a specific genomic insert of 8 kb was able to restore partially growth of the kre9∷HIS3 haploid strain. By using an Applied Biosystems 373A DNA sequencer, sequencing of this insert was initiated from both ends and was pursued until the KRE9 ORF was encountered, some 1,436 bp from the 5′ end of the complementing insert. The KRE9 region then was sequenced on both strands. A 1.6-kb DraI DNA fragment containing the complete CaKRE9 gene was subcloned from the original insert into the SmaI site and the filled XbaI site (treated with the Klenow fragment of DNA polymerase I) of YEp352 (see Fig. 3a) and was used in complementation studies (see Fig. 2).
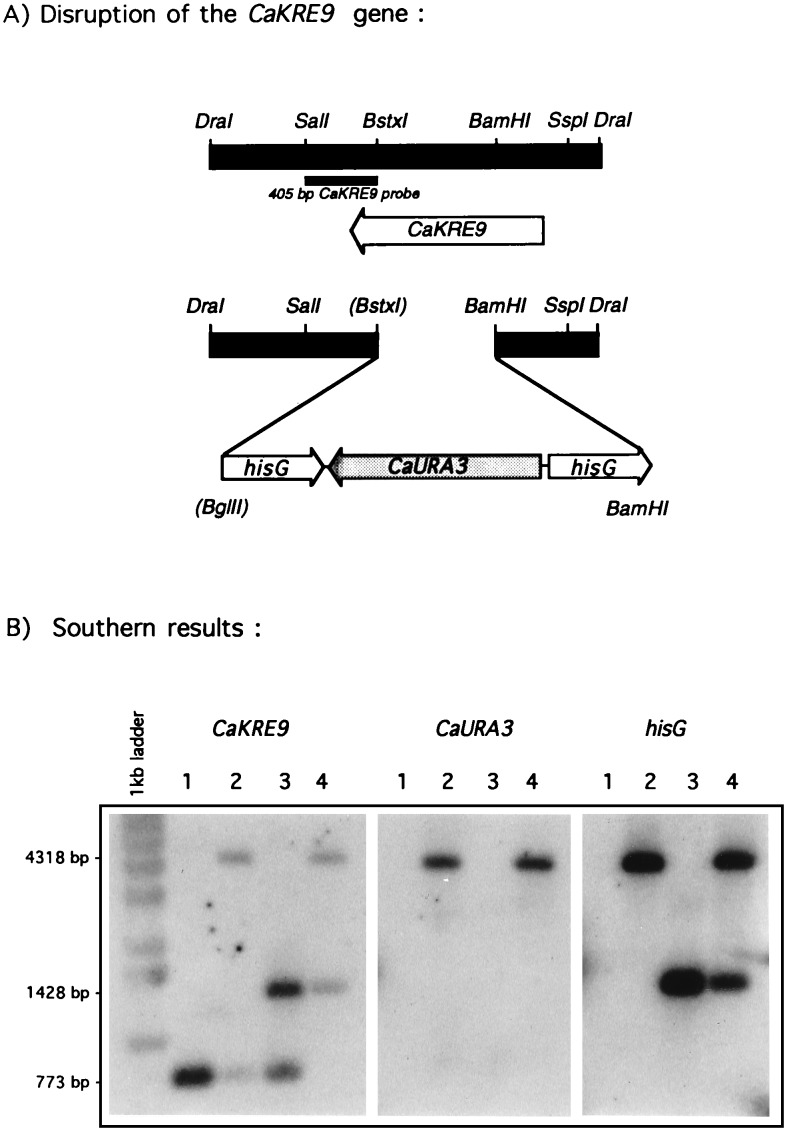
Figure 3.
Disruption of Candida albicans KRE9 gene. (A) Schematic representation of the disruption strategy (see Materials and Methods). (B) Results of the Southern blot verifications of the correct integration of the hisG-URA3-hisG disruption module into the CaKRE9 gene and proper CaURA3 excision after fluoroorotic acid treatment. Extracted genomic DNAs are from CAI4 wild-type cells (lane 1), CaKRE9/cakre9Δ∷hisG-URA3-hisG heterozygous mutant (lane 2), CaKRE9/cakre9Δ∷hisG heterozygous mutant obtained after fluoroorotic acid treatment (lane 3), and a cakre9Δ∷hisG/cakre9Δ:hisG-URA3-hisG homozygous null mutant that was grown on galactose (lane 4).
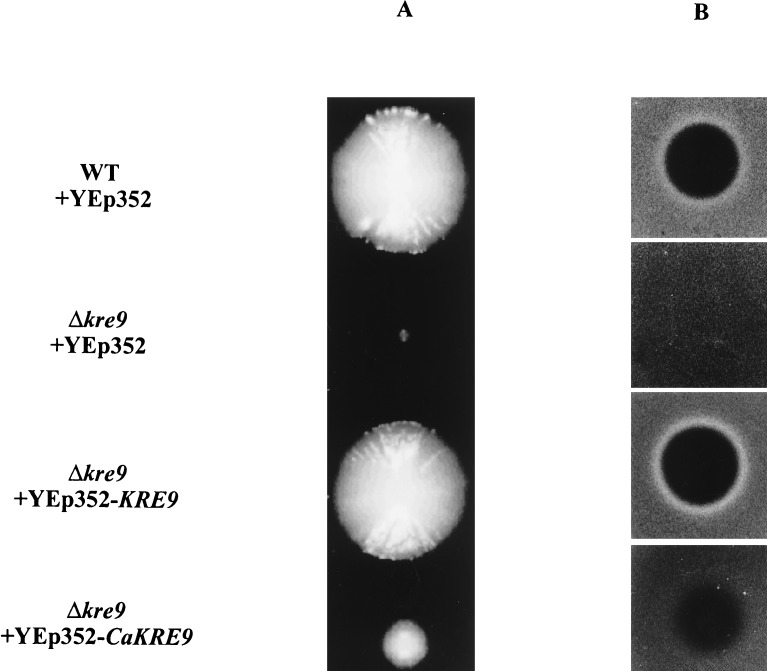
Figure 2.
Complementation of the growth (A) and killer resistance (B) phenotypes of the S. cerevisiae kre9Δ null mutant by the C. albicans KRE9 gene. Wild-type cells, a kre9Δ mutant, or the same mutant harboring KRE9 or CaKRE9 on multicopy vector YEp352 were scored for growth and for killer resistance in a seeded plate assay.
Disruption of the CaKRE9 Gene.
Gene disruption was performed by the URA blaster protocol by using the hisG-CaURA3-hisG module (20, 21). The CaKRE9 gene was disrupted by deleting a 485-bp BstxI-BamHI fragment of the ORF and replacing it with a 4.0-kb BglII/BamHI fragment carrying the hisG-URA3-hisG module from plasmid pCUB-6 (see Fig. 3a) (21). The sticky ends were treated with T4 DNA polymerase to accommodate the ligation. This disruption plasmid was digested by HindIII and KpnI, precipitated with ethanol and sodium acetate, and 100 μg of the 5.2-kb disruption fragment was transformed into CAI4 C. albicans cells (21) by the lithium acetate method (22, 23). Putative heterozygous disruptants were selected on minimal medium lacking uracil and containing either glucose or galactose as a carbon source. In preparation for a second round of gene disruption, the CaURA gene was excised by using a fluoroorotic acid selection (24). The second round of transformation was performed in the same way as the primary one. The accurate integration of the hisG-URA3-hisG cassette into the CaKRE9 gene and its excision from genomic DNA was verified by Southern hybridization by using three different probes: (i) a 405-bp fragment from C. albicans genomic DNA containing coding and 3′-flanking sequences of CaKRE9; (ii) a 783-bp DNA fragment obtained by PCR and covering the entire CaURA3 coding region; and (iii) a 898-bp fragment amplified by PCR that encompasses the whole of the Salmonella typhimurium hisG gene (see Fig. 3). All genomic DNAs were digested with the BamHI and SalI restriction enzymes.
β-1,6-Glucan Analysis of C. albicans CaKRE9 Mutants.
A specific rabbit anti-β-1,6-glucan antiserum was raised against BSA-coupled pustulan (25) and was used to detect antigen–antibody complexes by Western blotting of total cell protein extracts of different yeast strains as described below. Before use, the antiserum was affinity purified via chromatography by using pustulan-Sepharose 6B (Pharmacia), prepared as follows: pustulan was hydrolyzed (17 mg/ml in 0.1M NaOH, 1 h, 100°C) and subsequently was coupled to epoxy-activated Sepharose 6B as per the manufacturer’s instructions. For immunodetection of β-1,6-glucan, the following procedure was used. Exponentially growing cells were lysed by using glass beads and were standardized for total cellular protein. Subsequently, total cell lysate material representing equivalent amounts of protein were alkali extracted (0.75M NaOH, 1 h, 75°C). The alkali-soluble fractions then were spotted onto nitrocellulose, and immunoblots were treated in TBST buffer (10 mM Tris, pH 8.0/150 mM NaCl/0.05% Tween 20) containing 5% nonfat dried milk powder and subsequently were incubated with affinity purified rabbit anti-β-1,6-glucan antibodies in the same buffer. After antibody binding, membranes were washed in TBST, and a second antibody directed against rabbit Igs and conjugated with horseradish peroxidase was added. The blots again were washed, and whole cell alkali-soluble β-1,6-glucan was detected by using an enhanced chemiluminescence procedure. For cell wall analysis, the levels of cell wall alkali-insoluble β-1,3- and β-1,6-glucans were determined directly as described (18).
RESULTS
Isolation and Characterization of the CaKRE9 Gene.
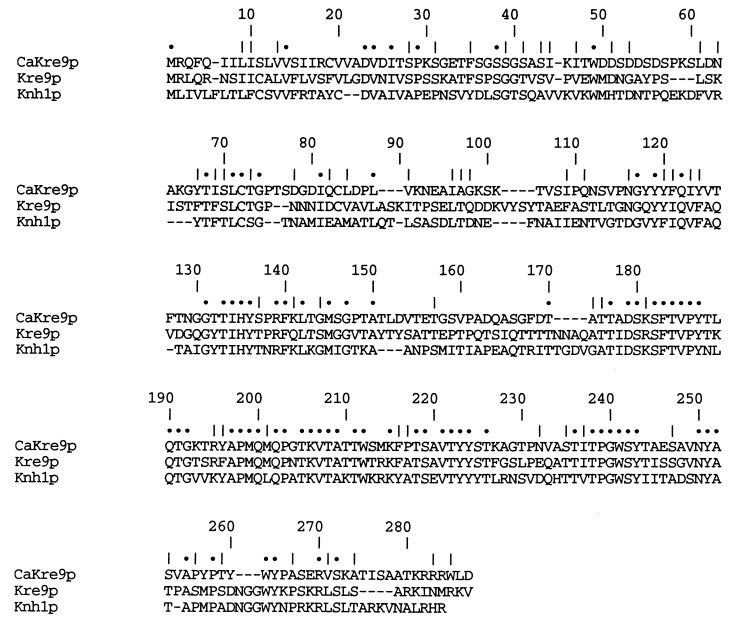
Using complementation of the S. cerevisiae kre9 mutant growth phenotype, we isolated a C. albicans gene that encodes a protein highly similar to the S. cerevisiae KRE9 gene product. Partial sequencing of the 8-kb fragment containing the CaKRE9 gene revealed an ORF of 813 bp (for details, see Materials and Methods) encoding a predicted 29-kDa secretory protein of 271 amino acid residues. As with Kre9p and Knh1p (17, 18), the hydrophobic N-terminal region of CaKre9p is a eukaryotic signal sequence, with the most likely cleavage site occurring at the peptide bond between amino acid residues 21 and 22. CaKre9p shares 43% overall amino acid identity with Kre9p and 32% with Knh1p (see Fig. 1). The most conserved region between the three proteins encompasses a large part of the central region and most of the C-terminal portion, with the N-terminal portion being largely unique to each protein. Kre9p, Knh1p, and CaKre9p share a high proportion of serine and threonine residues (26%), potential sites for O-glycosylation, a modification known to occur on Kre9p and Knh1p and characteristic of many yeast cell surface proteins. In addition, all three proteins have lysine and arginine rich C termini and lack potential N-linked glycosylation sites. The hydropathy profiles of all three proteins are similar (data not shown), which implies that they possess a similar secondary structure and which is consistent with their sharing related functions in complementing S. cerevisiae kre9 mutants (ref. 18 and see below).
Figure 1.
Comparison of the sequence of Kre9p from C. albicans and Kre9p and Knh1p from S. cerevisiae. The amino acid residues are shown in single-letter amino acid code. Sequences were aligned with gaps to maximize homology. Dots represent a perfect match between all three sequences whereas a vertical slash indicates a conservative substitution.
The ability of CaKre9p to function in S. cerevisiae was assessed by measuring its ability to restore the growth and killer toxin sensitivity of a kre9 null mutant (17). The YEp352-based cloned CaKRE9 gene from C. albicans genomic DNA was transformed into a diploid strain of S. cerevisiae heterozygous for a kre9∷HIS3 deletion. This diploid was sporulated, and a haploid kre9∷HIS3 strain containing a plasmid-based copy of CaKRE9 was obtained from spore progeny after tetrad dissection. As can be seen in Fig. 2a, a strain harboring the CaKRE9 gene grows at a slower rate than a wild-type strain or the mutant strain with a copy of the S. cerevisiae KRE9 but significantly faster than the kre9 null mutant alone, which displays a severe impairment of growth phenotype. The haploid S. cerevisiae kre9 strain carrying the CaKRE9 also was tested for killer toxin sensitivity (Fig. 2b). K1 killer yeast strains secrete a small pore-forming toxin that requires an intact cell wall receptor for function (26). ScKRE9 null mutants show an 80% reduction in the level of β-1,6-glucan, rendering the toxin receptor inactive and leading to killer resistance (17). The killer resistance phenotype of the kre9 mutant allowed a test of suppression by CaKre9p. Overexpression of CaKRE9 in the S. cerevisiae haploid strain carrying a disrupted copy of ScKRE9 partially suppressed the killer resistance phenotype (Fig. 2b). These results imply that ScKre9p and CaKre9p both play related roles in β-1,6-glucan assembly in S. cerevisiae and C. albicans.
Chromosomal Disruption of the CaKRE9 Gene.
In a first round of transformation where transformants were selected on glucose containing plates, Southern blotting results from transformant’s DNA revealed that the hisG-URA3-hisG module (see Materials and Methods) correctly integrated into the C. albicans KRE9 gene (Fig. 3). When genomic DNA of putative heterozygous CaKRE9 disruptions was digested with the SalI and BamHI restriction enzymes and was probed with the CaKRE9 405-bp SalI–BstXI DNA fragment along with the hisG and the CaURA3 probes, two expected bands could be detected (see Fig. 3b, lanes 1 and 2, for representative results): a 773-bp band corresponding to the wild-type gene that could only be detected by the CaKRE9 probe and a 4,318-bp diagnostic band, revealed by all three probes, indicating successful disruption of one copy of the CaKRE9 gene. After removal of the CaURA3 by using fluoroorotic acid, the 773-bp wild-type band could still be visualized, but the disrupted band from which the CaURA3 was excised shifted to an anticipated 1,428-bp when probed with the CaKRE9 and hisG probes (see Fig. 3b, lane 3).
To test whether the CaKRE9 gene is essential in C. albicans, a second round of disruption was undertaken in the heterozygous strain where one copy of CaKRE9 was disrupted and the CaURA3 gene was eliminated. In S. cerevisiae, kre9 mutants grow very poorly on glucose and considerably faster on galactose. This is partly because, in this organism, KRE9 has a functional homologue, KNH1. Transcription of the KNH1 gene is normally low in wild-type cells grown on glucose but increases ≈5-fold in galactose grown cells, where it partially compensates for the loss of Kre9p and allows partial suppression of the growth defect (18).
To avoid such a possible growth problem in a C. albicans kre9 disruptant when grown on glucose, the second round transformants were obtained on both galactose and glucose. The population of transformants obtained on galactose was heterogeneous with large and small sized colonies occurring (data not shown). To test for a possible carbon source dependence, 26 of these different sized colonies were plated from galactose to glucose. Among the smaller colonies, eight did not grow on glucose, suggesting they might be homozygous disruptants. Southern blot hybridizations were performed on all eight transformants, and they were shown to be homozygous disruptants for the CaKRE9 locus: One copy corresponded to the disrupted gene in which the C. albicans URA3 gene has been removed (1,428 bp), and the second represented the inactivation of the remaining wild-type copy by the hisG-URA3-hisG module (4,318 bp; see Fig. 3b, lane 4 for representative results). Thirty-two Ura+ colonies that were able to grow on glucose also were analyzed by Southern blot hybridization by using the three different probes, and only yeast cells heterozygous at the CaKRE9 locus could be found (data not shown). Thus, a homozygous disruption of kre9 in C. albicans is lethal when grown on glucose but can grow slowly when galactose is the carbon source.
The ability of the homozygous cakre9 disruptant to grow on carbon sources other than galactose was assessed. The homozygous cakre9Δ∷hisG/cakre9Δ∷hisG-URA3-hisG disruptant strain grew at or below the rate of growth on galactose on sucrose, mannose, maltose, fructose, sorbitol, and glycerol as carbon sources (data not shown). To test whether the lethality of the homozygous disruptant on glucose was caused by its inability to sense or use glucose, homozygous disruptant cells were scored for growth in the presence of glucose plus another of the above carbon sources (data not shown). Under such conditions, the homozygous cells could not grow, indicating that the lethality occurs when glucose is present. To extend these results, we examined the response of homozygous cakre9 disruptants over a range of glucose concentrations. Compared with wild-type cells, the homozygous mutant failed to grow at glucose concentrations from 1.0% to 0.1%–0.05%, with these lower concentrations approximating the glucose level in human serum (data not shown).
β-1,6-Glucan Analysis of C. albicans kre9 Mutants.
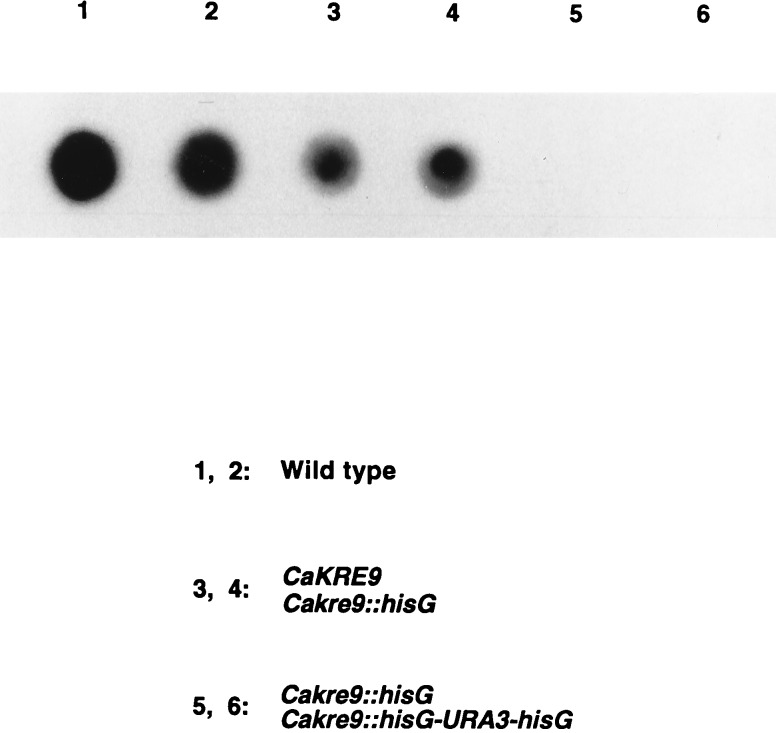
Whole cell alkali-soluble β-1,6-glucan levels in wild-type and cakre9 mutants were measured by using an affinity purified anti-β-1,6-glucan antibody (see Materials and Methods). Wild-type yeast grown on glucose and galactose yielded a strong β-1,6-glucan signal on either carbon source (see Fig. 4 for results on galactose). Quantitation of alkali-soluble β-1,6-glucan levels revealed that ≈20% of the amount observed in wild-type cells was present in the heterozygous CaKRE9/cakre9Δ whereas no β-1,6-glucan could be detected in extracts from a C. albicans homozygous cakre9Δ disruptant grown on galactose (Fig. 4). To extend these immunological results, determinations of the levels of cell wall alkali-insoluble glucans were made on the different strains. As seen in Table 1, a cakre9Δ heterozygous disruptant appears to possess an ≈28% decrease in the level of alkali-insoluble β-1,6-glucan compared with a wild type whereas in homozygous disruptant cells, no β-1,6-glucan could be measured, demonstrating that CaKre9p is required for cell wall β-1,6-glucan biosynthesis.
Figure 4.
Detection of cell wall β-1,6-glucan levels in wild-type and cakre9Δ mutants grown by using galactose as the sole carbon source. A specific anti-β-1,6-glucan antiserum was used to detect whole cell alkali-soluble β-1,6-glucan of different C. albicans strains by Western blotting followed by an enhanced chemiluminescence procedure (see Materials and Methods).
Table 1.
Cell wall polymer levels measured* in different C. albicans strains
| Genotype | β-1,6-Glucan† | β-1,3-Glucan + β-1,6-Glucan† |
|---|---|---|
| Wild-type | 235 ± 15 | 339 ± 17 |
| Heterozygous cakre9Δ | 169 ± 19 | 328 ± 17 |
| Homozygous cakre9Δ‡ | 0§ | 377 ± 4 |
| Homozygous cakre9Δ‡ | 0§ | 375 ± 21 |
Measured as micrograms of alkali-insoluble glucan per milligram dry weight cell wall, as described in Materials and Methods.
Values depict mean ±1 SD (n = 3).
These constitute two independent homozygous disruptants.
Values were found to be <1 μg.
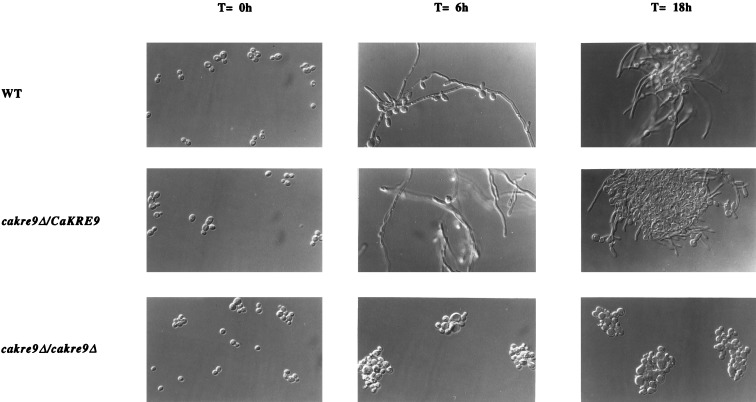
Hyphal Growth of cakre9Δ Mutants.
In S. cerevisiae, kre9 mutants have morphological defects, bud randomly, and fail to form mating projections with α-mating pheromone (17), so the possible role of CaKRE9 in hyphal formation was investigated. Germ tube formation was induced and hyphae formed when wild-type and heterozygous cakre9Δ disruptant cells were grown in presence of galactose and fetal bovine serum (27). However, no germ tube formation was observed in the cakre9Δ homozygous disruptant under the same conditions (Fig. 5).
Figure 5.
Induction of hyphal formation of different C. albicans strains. CAI4 wild-type, heterozygous mutant, and homozygous mutant cells were induced to form hyphae in presence of galactose and 20% fetal bovine serum at 37°C for up to 18 h. Cells were visualized by Nomarski optics.
DISCUSSION
CaKre9p is similar to Kre9p and Knh1p because it is predicted to possess an amino-terminal signal sequence and is rich in serine and threonine residues that could be O-modified (17, 18). Even though the proteins are very similar, the complementation of the growth and toxin sensitivity phenotypes of a S. cerevisiae KRE9 mutant by CaKRE9 in high copy number was partial. This incomplete complementation may be because the C. albicans promoter of CaKRE9 may not function correctly in S. cerevisiae. The isolation in C. albicans of a partially functional homologue of the S. cerevisiae KRE9/KNH1 pair strongly suggests that CaKre9p and Kre9p/Knh1p play similar roles in β-1,6-glucan synthesis or assembly in both species. Comparative studies with C. albicans have so far identified and characterized four genes, KRE1, SKN1, KRE6, and KRE9, involved in β-1,6-glucosylation based on their relatedness to those in S. cerevisiae, indicating that synthesis of this polymer is conserved functionally and is essential for the growth of C. albicans (refs. 16 and 19 and this work), indicating that both yeast species likely share similar sets of genes for the production of this glucan.
Results presented here establish that CaKre9p is required for synthesis of β-1,6-glucan in C. albicans. When one chromosomal copy is deleted, as found in the heterozygote, the levels of β-1,6-glucan are reduced. The anti-β-1,6-glucan antibody detected ≈20% of the wild-type levels of the alkali-soluble polymer in the heterozygous cakre9Δ. An enzymatic digestion procedure that measures β-1,3-glucanase-resistant, alkali-insoluble glucan detected a value of β-1,6-glucan of 70% of the wild-type level in such a heterozygous cakre9Δ strain. These differences in the levels of the alkali-soluble and alkali-insoluble fractions of the β-1,6-glucans suggest that the majority of the residual β-1,6-glucans in this heterozygous mutant is crosslinked to other polymers and is alkali-insoluble. Both evaluation methods, however, indicate that the homozygous disruptant lacks any detectable β-1,6-glucan polymer.
One explanation for the observation that a homozygous disruptant of CaKRE9 can grow when a carbon source other than glucose is used is that a second functionally homologous gene may be present in the C. albicans genome. However, in contrast to the situation observed in S. cerevisiae, a homozygous cakre9Δ null mutant has no detectable β-1,6-glucan whereas the kre9Δ null mutant retains 20% of the wild-type levels of this polymer, an amount thought to be contributed by the homologous KNH1 gene (18). Thus, if there were another KRE9/KNH1 homologue in C. albicans, we would expect to find a certain amount of cell wall β-1,6-glucan when the homozygous cakre9Δ was grown on galactose, yet we do not. Consistent with this, efforts to find a second C. albicans gene have so far been unsuccessful. Low stringency Southern blots, PCR approaches using degenerate primers, and a functional complementation approach with galactose as a carbon source have not yielded a second gene. It remains to be determined whether a duplicated copy of CaKRE9 is present in C. albicans, but the complete absence of β-1,6-glucan in the homozygous cakre9Δ deletion suggests that this is not likely to be the case.
We offer two kinds of possible explanation for the galactose-specific survival of homozygous cakre9 null mutants. The first is that some compensatory mechanism exists in galactose-grown cells that does not occur when glucose is present, perhaps through glucose repression. For example, in the absence of glucose, a lack of β-1,6-glucan might be compensated for by an increase in levels of β-1,3-glucan, chitin, or mannoproteins or in altered crosslinking of these polymers. In a contrasting model, glucose-grown cells respond to a wall defect by attempting to compensate in a way that has lethal consequences in a cakre9 mutant. For example, cells may overproduce chitin or crosslink it to other polymers to such an extent that it prevents growth of the cell wall. Such a lethal compensatory pathway would be glucose-specific and would not occur when cells are grown on galactose.
A number of attributes suggest that the KRE9 gene of C. albicans is a potentially useful antifungal target. CaKRE9 is involved in a fungal specific process, cell wall β-1,6-glucan synthesis. The gene is not found in the sequenced bacterial (28, 29) and archaebacterial genomes (30) and appears to be absent from metazoans, with no known homologues present in the C. elegans, mouse, and Homo sapiens databases. CaKRE9 is an essential gene when grown on glucose or when glucose is present at levels comparable to those found in human serum. Thus, a C. albicans strain in which CaKre9p would be inactivated/inhibited by a drug should be unable to grow in the presence of glucose. In the event that, in a human host, an alternative carbon source would be present in the absence of glucose, a CaKre9p inhibited C. albicans would, based on our laboratory studies, be expected to grow poorly and be unable to form hyphae. This inability of cakre9 mutants to form germ tubes or hyphae extends the morphological budding and mating projection defects seen in S. cerevisiae kre9 mutants (17); thus, Kre9p and CaKre9p or the presence of β-1,6-glucan may be necessary for implementing normal, polarized cell wall growth in buds and hyphae.
The exact secretory pathway location of the KRE9 gene product is not known, but the available data suggest that it functions extracellularly at the cell surface (17, 18). If this is the case, an inhibitor of CaKre9p need not enter the fungal cell to be effective, thus reducing the risk of resistance occurring through reduced uptake or increased efflux of the drug by membrane pumps.
Acknowledgments
We thank Ying Wang and Tiziano di Paulo for their assistance with DNA sequencing and Shigehisa Nagahashi and Peter Dijkgraaf for discussions and for critically reading the manuscript. This work was supported by an Operating Grant from the Natural Sciences and Engineering Research Council of Canada. H.B. is a Canadian Pacific Professor.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF069763).
References
- 1.Pfaller, M. A. (1995) J. Hosp. Infect. 30, Suppl., 329–338. [DOI] [PubMed]
- 2.Pfaller, M. A. (1996) Clin. Infect. Dis. 22, Suppl. 2, S89–S94. [DOI] [PubMed]
- 3.Odds F C. Int J Antimicrob Agents. 1996;6:141–144. doi: 10.1016/0924-8579(95)00049-6. [DOI] [PubMed] [Google Scholar]
- 4.Wingard J R. Clin Infect Dis. 1995;20:115–125. doi: 10.1093/clinids/20.1.115. [DOI] [PubMed] [Google Scholar]
- 5.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, et al. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen M H, Peacock J E, Morris A J, Tanner D C, Nguyen M L, Snydman D R, Wagener M M, Rinaldi M G, Yu V L. Am J Med. 1996;100:617–623. doi: 10.1016/s0002-9343(95)00010-0. [DOI] [PubMed] [Google Scholar]
- 7.Odds F C. Int J Antimicrob Agents. 1996;6:145–147. doi: 10.1016/0924-8579(95)00048-8. [DOI] [PubMed] [Google Scholar]
- 8.Klis F. Yeast. 1994;10:851–869. doi: 10.1002/yea.320100702. [DOI] [PubMed] [Google Scholar]
- 9.Cid V J, Duran A, del Rey F, Snyder M P, Nombela C, Sanchez M. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orlean P. In: The Molecular and Cellular Biology of the Yeast Saccharomyces. Pringle J R, Broach J R, Jones E W, editors. Vol. 3. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 229–362. [Google Scholar]
- 11.Kollar R, Reinhold B B, Petrakova E, Yeh H J C, Ashwell G, Drgonova J, Kapteyn J C, Klis F M, Cabib E. J Biol Chem. 1997;272:17762–17775. doi: 10.1074/jbc.272.28.17762. [DOI] [PubMed] [Google Scholar]
- 12.Sietsma J H, Wessels J G. J Gen Microbiol. 1981;125:209–212. doi: 10.1099/00221287-125-1-209. [DOI] [PubMed] [Google Scholar]
- 13.Sietsma J H, Wessels J G. J Gen Microbiol. 1990;136:2261–2265. doi: 10.1099/00221287-136-11-2261. [DOI] [PubMed] [Google Scholar]
- 14.Montijn R C, Van Wolven P, De Hoog S, Klis F M. Microbiology. 1997;143:1673–1680. doi: 10.1099/00221287-143-5-1673. [DOI] [PubMed] [Google Scholar]
- 15.Gopal P K, Maxwell G, Sullivan P A. J Gen Microbiol. 1984;130:3295–3301. doi: 10.1099/00221287-130-12-3295. [DOI] [PubMed] [Google Scholar]
- 16.Mio T, Yamada-Okabe T, Yabe T, Nakajima T, Arisawa M, Yamada-Okabe H. J Bacteriol. 1997;179:2363–2372. doi: 10.1128/jb.179.7.2363-2372.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown J L, Bussey H. Mol Cell Biol. 1993;13:6346–6356. doi: 10.1128/mcb.13.10.6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dijkgraaf G J, Brown J L, Bussey H. Yeast. 1996;12:683–692. doi: 10.1002/(SICI)1097-0061(19960615)12:7%3C683::AID-YEA959%3E3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 19.Boone C, Sdicu A M, Laroche M, Bussey H. J Bacteriol. 1991;173:6859–6864. doi: 10.1128/jb.173.21.6859-6864.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alani E, Cao L, Kleckner N. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fonzi W A, Irwin M Y. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanglard D, Ischer F, Monod M, Bille J. Antimicrob Agents Chemother. 1996;40:2300–2305. doi: 10.1128/aac.40.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gietz R D, Schiestl R H, Willems A R, Woods R A. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 24.Sikorski R S, Boeke J D. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- 25.Montijn R C, van Rinsum J, van Schagen F A, Klis F M. J Biol Chem. 1994;269:19338–19342. [PubMed] [Google Scholar]
- 26.Bussey H. Mol Microbiol. 1991;5:2339–2343. doi: 10.1111/j.1365-2958.1991.tb02079.x. [DOI] [PubMed] [Google Scholar]
- 27.Bulawa C E, Miller D W, Henry L K, Becker J M. Proc Natl Acad Sci USA. 1995;92:10570–10574. doi: 10.1073/pnas.92.23.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blattner F R, Plunkett G, III, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 29.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 30.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]