Abstract
Mutants separately resistant to novobiocin, coumermycin, nalidixic acid, and oxolinic acid contained gyrase activity as measured in vitro that was resistant to the antibiotics, indicating that the mutations represented structural alterations of the enzyme. One Novr mutant contained an altered B subunit of the enzyme, as judged by the ability of a plasmid, pNov1, containing the mutation to complement a temperature-sensitive gyrase B mutation in Escherichia coli and to cause novobiocin resistance in that strain. Three other Novr mutations did not confer antibiotic resistance to the gyrase but appeared to increase the amount of active enzyme in the cell. One of these, novB1, could only act in cis, whereas a new mutation, novC, could act in trans. An RNA polymerase mutation partially substituted for the novB1 mutation, suggesting that novB1 may be a mutation in a promoter region for the B subunit gene. Growth responses of strains containing various combinations of mutations on plasmids or on the chromosome indicated that low-level resistance to novobiocin or coumermycin may have resulted from multiple copies of wild-type genes coding for the gyrase B subunit, whereas high-level resistance required a structural change in the gyrase B gene and was also dependent on alteration in a regulatory region. When there was mismatch at the novB locus, with the novB1 mutation either on a plasmid or the chromosome, and the corresponding wild-type gene present in trans, chromosome to plasmid recombination during transformation was much higher than when the genes matched, probably because plasmid to chromosome recombination, eliminating the plasmid, was inhibited by the mismatch.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beattie K. L., Setlow J. K. Repair of ultraviolet-irradiated transforming deoxyribonucleic acid in Haemophilus influenzae. J Bacteriol. 1970 Mar;101(3):808–812. doi: 10.1128/jb.101.3.808-812.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boling M. E., Allison D. P., Setlow J. K. Bacteriophage of Haemophilus influenzae. 3. Morphology, DNA homology, and immunity properties of HPlcl, S2, and the defective bacteriophage from strain Rd. J Virol. 1973 Apr;11(4):585–591. doi: 10.1128/jvi.11.4.585-591.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
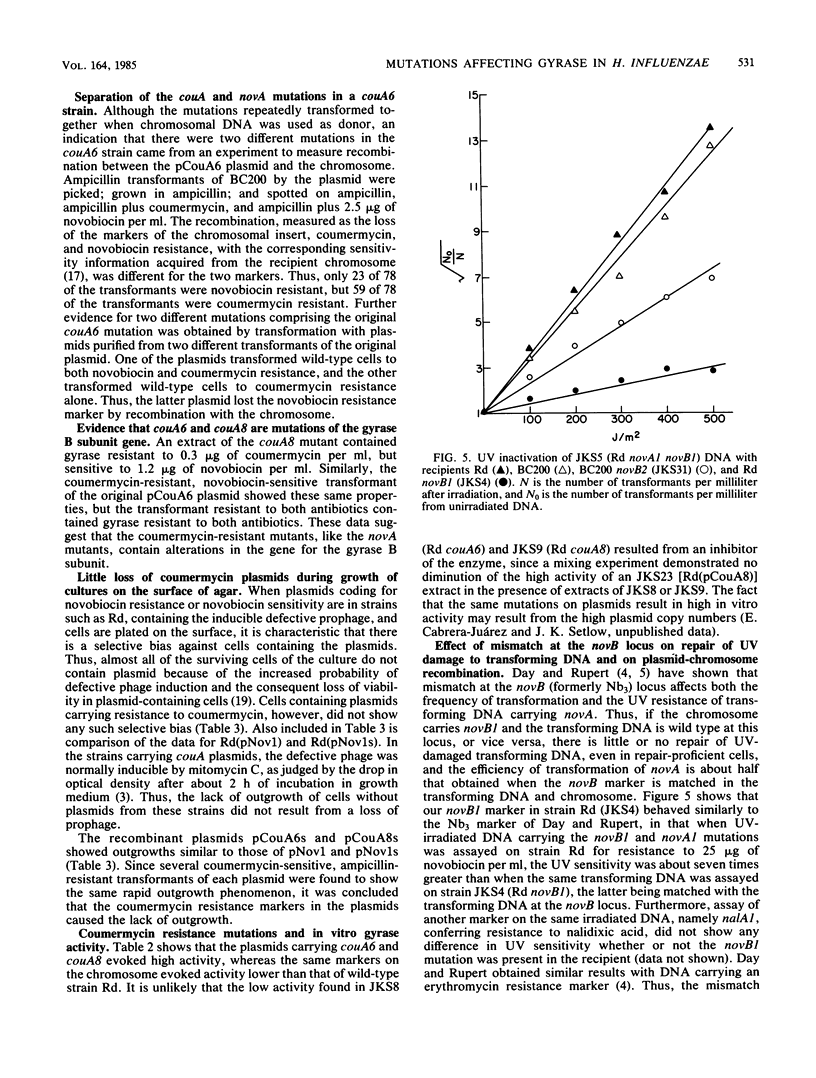
- Day R. S., 3rd, Rupert C. S. Ultraviolet sensitivity of Haemophilus influenzae transforming DNA. I. Effects of genetic mismatch and target size. Mutat Res. 1971 Mar;11(3):293–311. [PubMed] [Google Scholar]
- Day R. S., 3rd, Rupert C. S. Ultraviolet sensitivity of Haemophilus influenzae transforming DNA. II. A reextraction study of integration and repair. Mutat Res. 1971 Mar;11(3):313–326. [PubMed] [Google Scholar]
- Deneer H. G., Slaney L., Maclean I. W., Albritton W. L. Mobilization of nonconjugative antibiotic resistance plasmids in Haemophilus ducreyi. J Bacteriol. 1982 Feb;149(2):726–732. doi: 10.1128/jb.149.2.726-732.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOODGAL S. H. Studies on transformations of Hemophilus influenzae. IV. Linked and unlinked transformations. J Gen Physiol. 1961 Nov;45:205–228. doi: 10.1085/jgp.45.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., O'Dea M. H., Itoh T., Tomizawa J. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R., Gellert M. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell. 1983 Aug;34(1):105–113. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- Notani N. K., Setlow J. K., McCarthy D., Clayton N. L. Transformation of Haemophilus influenzae by plasmid RSF0885. J Bacteriol. 1981 Dec;148(3):812–816. doi: 10.1128/jb.148.3.812-816.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peebles C. L., Higgins N. P., Kreuzer K. N., Morrison A., Brown P. O., Sugino A., Cozzarelli N. R. Structure and activities of Escherichia coli DNA gyrase. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):41–52. doi: 10.1101/sqb.1979.043.01.008. [DOI] [PubMed] [Google Scholar]
- Setlow J. K., Boling M. E., Allison D. P., Beattie K. L. Relationship between prophage induction and transformation in Haemophilus influenzae. J Bacteriol. 1973 Jul;115(1):153–161. doi: 10.1128/jb.115.1.153-161.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow J. K., Boling M. E., Beattie K. L., Kimball R. F. A complex of recombination and repair genes in Haemophilus influenzae. J Mol Biol. 1972 Jul 21;68(2):361–378. doi: 10.1016/0022-2836(72)90218-5. [DOI] [PubMed] [Google Scholar]
- Setlow J. K., Brown D. C., Boling M. E., Mattingly A., Gordon M. P. Repair of deoxyribonucleic acid in Haemophilus influenzae. I. X-ray sensitivity of ultraviolet-sensitive mutants and their behavior as hosts to ultraviolet-irradiated bacteriophage and transforming deoxyribonucleic acid. J Bacteriol. 1968 Feb;95(2):546–558. doi: 10.1128/jb.95.2.546-558.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow J. K., Cabrera-Juárez E., Griffin K. Mechanism of acquisition of chromosomal markers by plasmids in Haemophilus influenzae. J Bacteriol. 1984 Nov;160(2):662–667. doi: 10.1128/jb.160.2.662-667.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow J. K., Notani N. K., McCarthy D., Clayton N. L. Transformation of Haemophilus influenzae by plasmid RSF0885 containing a cloned segment of chromosomal deoxyribonucleic acid. J Bacteriol. 1981 Dec;148(3):804–811. doi: 10.1128/jb.148.3.804-811.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow J. K., Spikes D., Ledbetter M. Loss of plasmids containing cloned inserts coding for novobiocin resistance or novobiocin sensitivity in Haemophilus influenzae. J Bacteriol. 1984 Jun;158(3):872–877. doi: 10.1128/jb.158.3.872-877.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhart W. L., Herriott R. M. Fate of recipient deoxyribonucleic acid during transformation in Haemophilus influenzae. J Bacteriol. 1968 Nov;96(5):1718–1724. doi: 10.1128/jb.96.5.1718-1724.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A., Higgins N. P., Brown P. O., Peebles C. L., Cozzarelli N. R. Energy coupling in DNA gyrase and the mechanism of action of novobiocin. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4838–4842. doi: 10.1073/pnas.75.10.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A., Peebles C. L., Kreuzer K. N., Cozzarelli N. R. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4767–4771. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland J. C., Monteleone D. C., Trunk J., Ciarrocchi G. Two-dimensional, computer-controlled film scanner: quantitation of fluorescence from ethidium bromide-stained DNA gels. Anal Biochem. 1984 Jun;139(2):390–399. doi: 10.1016/0003-2697(84)90023-x. [DOI] [PubMed] [Google Scholar]
- Voll M. J., Goodgal S. H. Loss of activity of transforming deoxyribonucleic acid after uptake by Haemophilus influenzae. J Bacteriol. 1965 Oct;90(4):873–883. doi: 10.1128/jb.90.4.873-883.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]



