Abstract
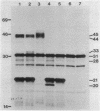
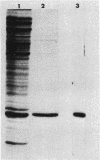
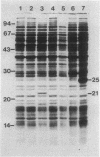
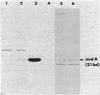
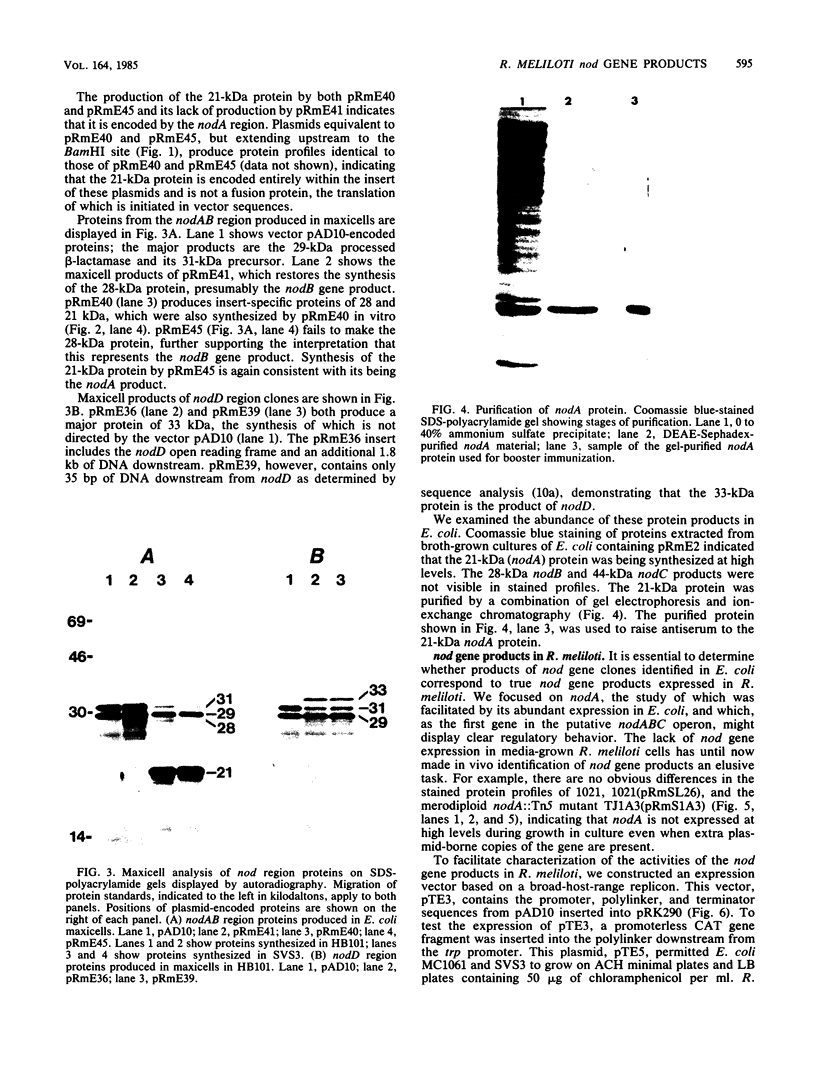
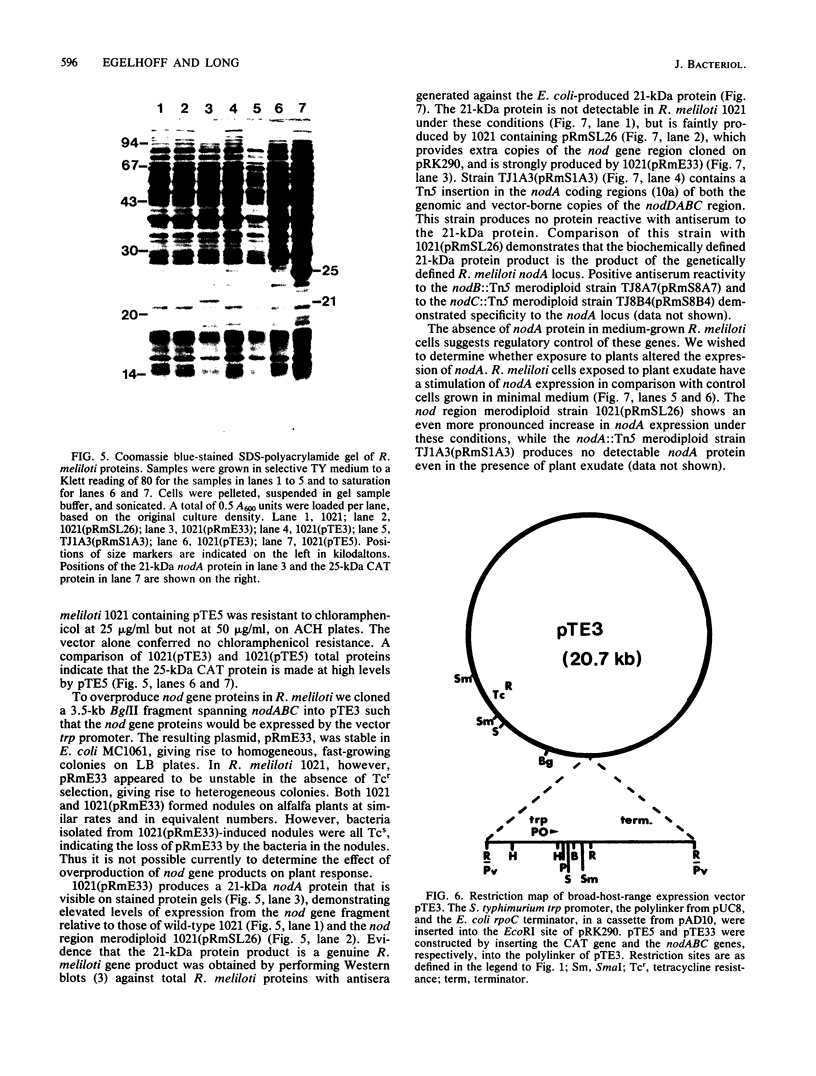
A set of conserved, or common, bacterial nodulation (nod) loci is required for host plant infection by Rhizobium meliloti and other Rhizobium species. Four such genes, nodDABC, have been indicated in R. meliloti 1021 by genetic analysis and DNA sequencing. An essential step toward understanding the function of these genes is to characterize their protein products. We used in vitro and maxicell Escherichia coli expression systems, together with gel electrophoresis and autoradiography, to detect proteins encoded by nodDABC. We facilitated expression of genes on these DNA fragments by inserting them downstream of the Salmonella typhimurium trp promoter, both in colE1 and incP plasmid-based vectors. Use of the incP trp promoter plasmid allowed overexpression of a nodABC gene fragment in R. meliloti. We found that nodA encodes a protein of 21 kilodaltons (kDa), and nodB encodes one of 28 kDa; the nodC product appears as two polypeptide bands at 44 and 45 kDa. Expression of the divergently read nodD yields a single polypeptide of 33 kDa. Whether these represent true Rhizobium gene products must be demonstrated by correlating these proteins with genetically defined Rhizobium loci. We purified the 21-kDa putative nodA protein product by gel electrophoresis, selective precipitation, and ion-exchange chromatography and generated antiserum to the purified gene product. This permitted the immunological demonstration that the 21-kDa protein is present in wild-type cells and in nodB- or nodC-defective strains, but is absent from nodA::Tn5 mutants, which confirms that the product expressed in E. coli is identical to that produced by R. meliloti nodA. Using antisera detection, we found that the level of nodA protein is increased by exposure of R. meliloti cells to plant exudate, indicating regulation of the bacterial nod genes by the plant host.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beringer J. E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974 Sep;84(1):188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- Burnette W. N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Bánfalvi Z., Sakanyan V., Koncz C., Kiss A., Dusha I., Kondorosi A. Location of nodulation and nitrogen fixation genes on a high molecular weight plasmid of R. meliloti. Mol Gen Genet. 1981;184(2):318–325. doi: 10.1007/BF00272925. [DOI] [PubMed] [Google Scholar]
- Chakravorty A. K., Zurkowski W., Shine J., Rolfe B. G. Symbiotic nitrogen fixation: molecular cloning of Rhizobium genes involved in exopolysaccharide synthesis and effective nodulation. J Mol Appl Genet. 1982;1(6):585–596. [PubMed] [Google Scholar]
- Close T. J., Rodriguez R. L. Construction and characterization of the chloramphenicol-resistance gene cartridge: a new approach to the transcriptional mapping of extrachromosomal elements. Gene. 1982 Dec;20(2):305–316. doi: 10.1016/0378-1119(82)90048-8. [DOI] [PubMed] [Google Scholar]
- Crouse G. F., Frischauf A., Lehrach H. An integrated and simplified approach to cloning into plasmids and single-stranded phages. Methods Enzymol. 1983;101:78–89. doi: 10.1016/0076-6879(83)01006-x. [DOI] [PubMed] [Google Scholar]
- Das A., Urbanowski J., Weissbach H., Nestor J., Yanofsky C. In vitro synthesis of the tryptophan operon leader peptides of Escherichia coli, Serratia marcescens, and Salmonella typhimurium. Proc Natl Acad Sci U S A. 1983 May;80(10):2879–2883. doi: 10.1073/pnas.80.10.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhoff T. T., Fisher R. F., Jacobs T. W., Mulligan J. T., Long S. R. Nucleotide sequence of Rhizobium meliloti 1021 nodulation genes: nodD is read divergently from nodABC. DNA. 1985 Jun;4(3):241–248. doi: 10.1089/dna.1985.4.241. [DOI] [PubMed] [Google Scholar]
- Finan T. M., Hirsch A. M., Leigh J. A., Johansen E., Kuldau G. A., Deegan S., Walker G. C., Signer E. R. Symbiotic mutants of Rhizobium meliloti that uncouple plant from bacterial differentiation. Cell. 1985 Apr;40(4):869–877. doi: 10.1016/0092-8674(85)90346-0. [DOI] [PubMed] [Google Scholar]
- Fisher R. F., Tu J. K., Long S. R. Conserved Nodulation Genes in Rhizobium meliloti and Rhizobium trifolii. Appl Environ Microbiol. 1985 Jun;49(6):1432–1435. doi: 10.1128/aem.49.6.1432-1435.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus R. P., Zurawski G., Yanofsky C. Structural and functional analysis of cloned deoxyribonucleic acid containing the trpR-thr region of the Escherichia coli chromosome. J Bacteriol. 1979 Oct;140(1):106–113. doi: 10.1128/jb.140.1.106-113.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch A. M., Drake D., Jacobs T. W., Long S. R. Nodules are induced on alfalfa roots by Agrobacterium tumefaciens and Rhizobium trifolii containing small segments of the Rhizobium meliloti nodulation region. J Bacteriol. 1985 Jan;161(1):223–230. doi: 10.1128/jb.161.1.223-230.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert R., Brusilow W. S., Gunsalus R. P., Klionsky D. J., Simoni R. D. Escherichia coli mutants defective in the uncH gene. J Bacteriol. 1983 Jan;153(1):416–422. doi: 10.1128/jb.153.1.416-422.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs T. W., Egelhoff T. T., Long S. R. Physical and genetic map of a Rhizobium meliloti nodulation gene region and nucleotide sequence of nodC. J Bacteriol. 1985 May;162(2):469–476. doi: 10.1128/jb.162.2.469-476.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lemaux P. G., Grossman A. Isolation and characterization of a gene for a major light-harvesting polypeptide from Cyanophora paradoxa. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4100–4104. doi: 10.1073/pnas.81.13.4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade H. M., Long S. R., Ruvkun G. B., Brown S. E., Ausubel F. M. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982 Jan;149(1):114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Normark S., Bergström S., Edlund T., Grundström T., Jaurin B., Lindberg F. P., Olsson O. Overlapping genes. Annu Rev Genet. 1983;17:499–525. doi: 10.1146/annurev.ge.17.120183.002435. [DOI] [PubMed] [Google Scholar]
- Rosenberg C., Boistard P., Dénarié J., Casse-Delbart F. Genes controlling early and late functions in symbiosis are located on a megaplasmid in Rhizobium meliloti. Mol Gen Genet. 1981;184(2):326–333. doi: 10.1007/BF00272926. [DOI] [PubMed] [Google Scholar]
- Rossen L., Johnston A. W., Downie J. A. DNA sequence of the Rhizobium leguminosarum nodulation genes nodAB and C required for root hair curling. Nucleic Acids Res. 1984 Dec 21;12(24):9497–9508. doi: 10.1093/nar/12.24.9497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp W. D., Wilde C. E., 3rd, Reno D. L., Howard-Flanders P. Exchanges between DNA strands in ultraviolet-irradiated Escherichia coli. J Mol Biol. 1971 Oct 14;61(1):25–44. doi: 10.1016/0022-2836(71)90204-x. [DOI] [PubMed] [Google Scholar]
- Sancar A., Wharton R. P., Seltzer S., Kacinski B. M., Clarke N. D., Rupp W. D. Identification of the uvrA gene product. J Mol Biol. 1981 May 5;148(1):45–62. doi: 10.1016/0022-2836(81)90234-5. [DOI] [PubMed] [Google Scholar]
- Schmidt J., John M., Kondorosi E., Kondorosi A., Wieneke U., Schröder G., Schröder J., Schell J. Mapping of the protein-coding regions of Rhizobium meliloti common nodulation genes. EMBO J. 1984 Aug;3(8):1705–1711. doi: 10.1002/j.1460-2075.1984.tb02035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Török I., Kondorosi E., Stepkowski T., Pósfai J., Kondorosi A. Nucleotide sequence of Rhizobium meliloti nodulation genes. Nucleic Acids Res. 1984 Dec 21;12(24):9509–9524. doi: 10.1093/nar/12.24.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Yanofsky C., Kelley R. L., Horn V. Repression is relieved before attenuation in the trp operon of Escherichia coli as tryptophan starvation becomes increasingly severe. J Bacteriol. 1984 Jun;158(3):1018–1024. doi: 10.1128/jb.158.3.1018-1024.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]