Abstract
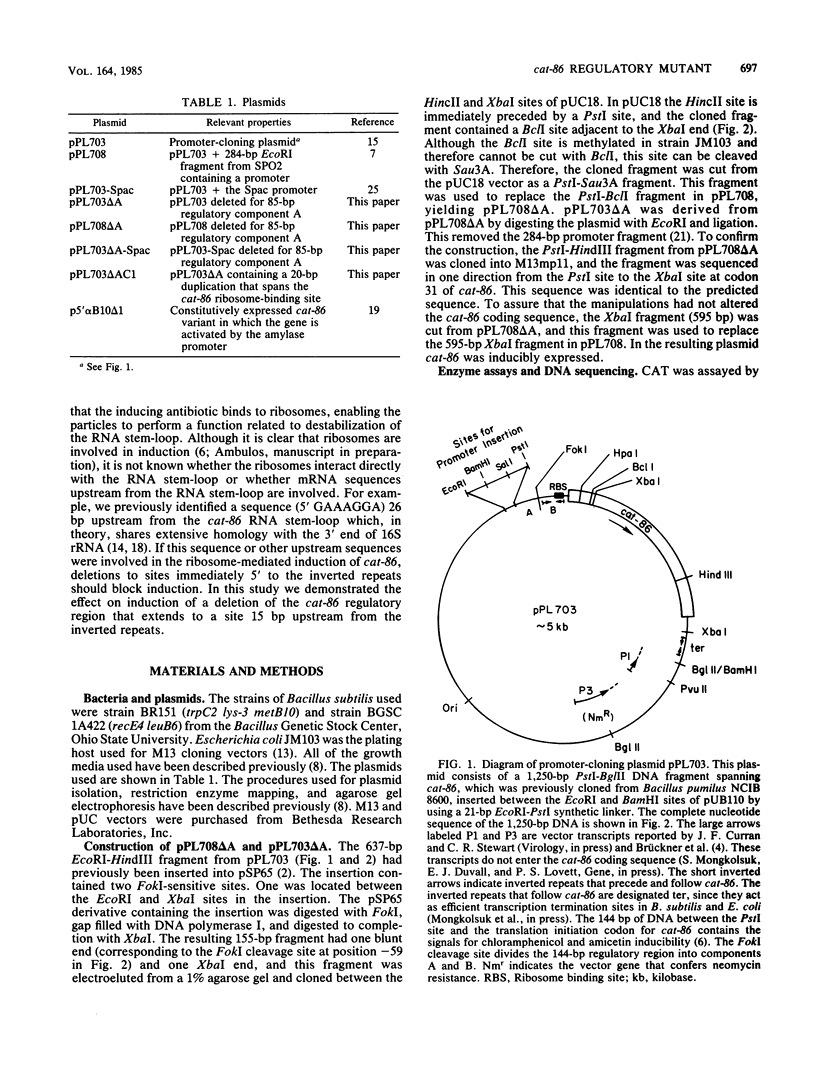
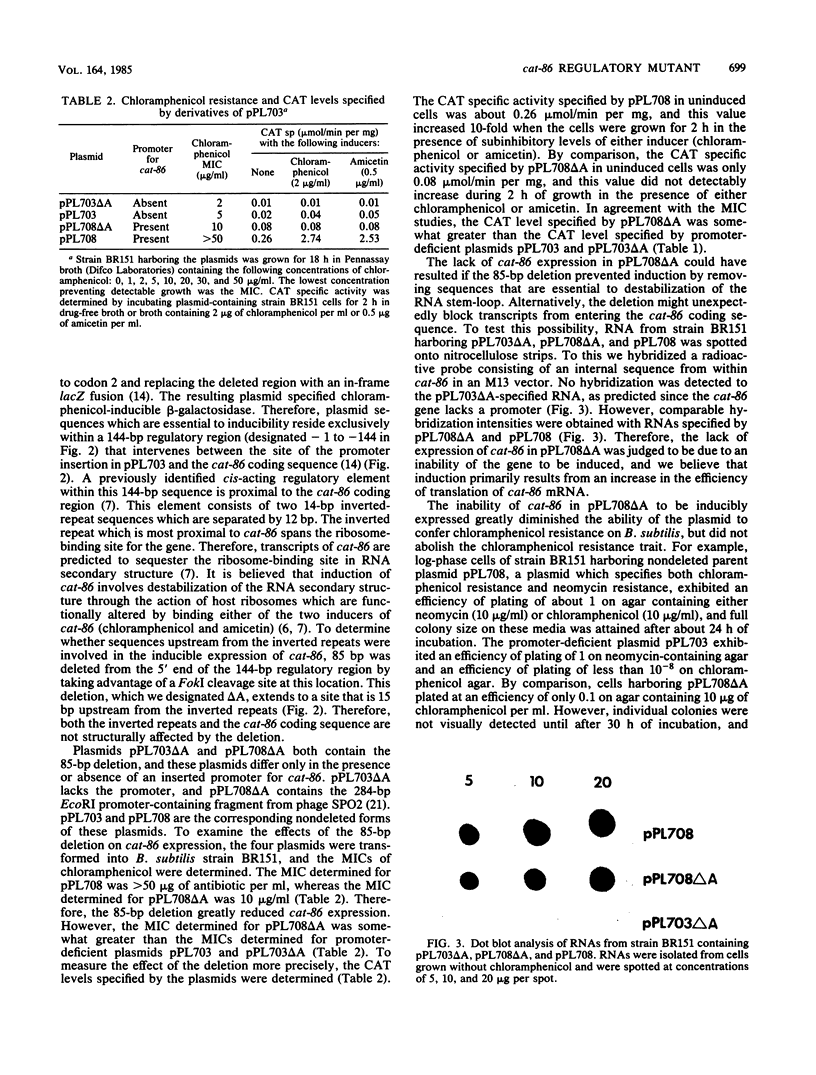
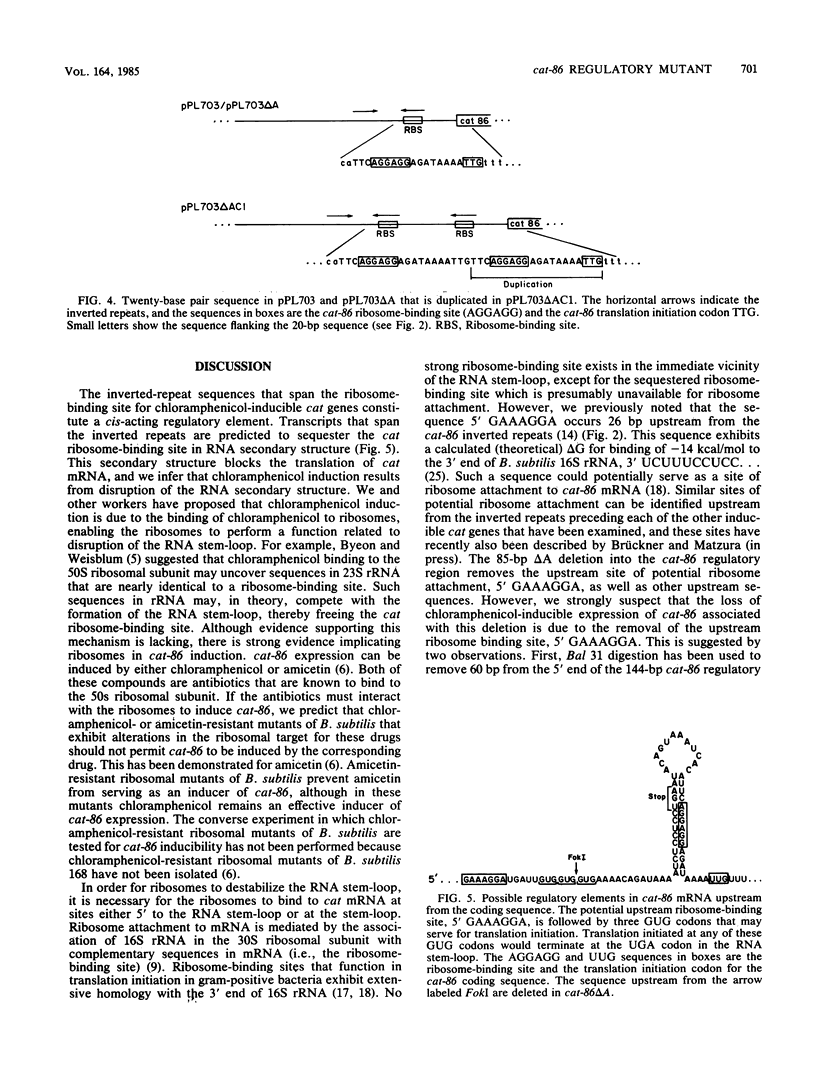
Sequences essential to the chloramphenicol-inducible expression of cat-86, a chloramphenicol acetyltransferase gene, reside in a 144-base pair (bp) regulatory region that intervenes between the cat-86 coding sequence and its promoter. A key regulatory element within the 144-bp segment consists of a pair of inverted-repeat sequences that immediately precede the cat-86 coding region and span the ribosome-binding site for the gene. Because of the location of the inverted repeats, cat-86 transcripts are predicted to sequester the ribosome-binding site in a stable RNA stem-loop structure which should block translation of cat-86 mRNA. Chloramphenicol induction of gene expression is believed to result from ribosome-mediated destabilization of the RNA stem-loop structure, which frees the cat-86 ribosome-binding site, thereby allowing translation. In this study we demonstrated that deletion of 85 bp from the 5' end of the 144-bp regulatory region abolishes inducible expression of cat-86, although the gene is transcribed. This deletion leaves intact both the inverted repeats and the cat-86 coding sequence, and the deletion mutation is not complementable. Therefore, inducible regulation of cat-86 requires the inverted repeats plus an upstream, cis-acting regulatory region. The cis-acting region is believed to control translation of cat-86 mRNA by its essential participation in chloramphenicol-induced opening of the RNA stem-loop. cat-86 deleted for the 85-bp regulatory region and therefore virtually unexpressed was used to select for mutations that restore expression to the gene. An analysis of one mutant plasmid showed that the cat-86 gene is constitutively expressed and that this results from a duplication of the DNA sequence that spans the ribosome-binding site. The duplication provides cat-86 with two ribosome-binding sites. One of these sites is predicted to be sequestered in an RNA stem-loop, and the other is not involved in RNA secondary structure.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambulos N. P., Jr, Chow J. H., Mongkolsuk S., Preis L. H., Vollmar W. R., 2nd, Lovett P. S. Constitutive variants of the pC194 cat gene exhibit DNA alterations in the vicinity of the ribosome binding site sequence. Gene. 1984 May;28(2):171–176. doi: 10.1016/0378-1119(84)90254-3. [DOI] [PubMed] [Google Scholar]
- Ambulos N. P., Jr, Mongkolsuk S., Lovett P. S. A transcription termination signal immediately precedes the coding sequence for the chloramphenicol-inducible plasmid gene cat-86. Mol Gen Genet. 1985;199(1):70–75. doi: 10.1007/BF00327512. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brückner R., Zyprian E., Matzura H. Expression of a chloramphenicol-resistance determinant carried on hybrid plasmids in gram-positive and gram-negative bacteria. Gene. 1984 Dec;32(1-2):151–160. doi: 10.1016/0378-1119(84)90043-x. [DOI] [PubMed] [Google Scholar]
- Byeon W. H., Weisblum B. Post-transcriptional regulation of chloramphenicol acetyl transferase. J Bacteriol. 1984 May;158(2):543–550. doi: 10.1128/jb.158.2.543-550.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall E. J., Mongkolsuk S., Kim U. J., Lovett P. S., Henkin T. M., Chambliss G. H. Induction of the chloramphenicol acetyltransferase gene cat-86 through the action of the ribosomal antibiotic amicetin: involvement of a Bacillus subtilis ribosomal component in cat induction. J Bacteriol. 1985 Feb;161(2):665–672. doi: 10.1128/jb.161.2.665-672.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall E. J., Williams D. M., Lovett P. S., Rudolph C., Vasantha N., Guyer M. Chloramphenicol-inducible gene expression in Bacillus subtilis. Gene. 1983 Oct;24(2-3):171–177. doi: 10.1016/0378-1119(83)90077-x. [DOI] [PubMed] [Google Scholar]
- Duvall E. J., Williams D. M., Mongkolsuk S., Lovett P. S. Regulatory regions that control expression of two chloramphenicol-inducible cat genes cloned in Bacillus subtilis. J Bacteriol. 1984 Jun;158(3):784–790. doi: 10.1128/jb.158.3.784-790.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- Harwood C. R., Williams D. M., Lovett P. S. Nucleotide sequence of a Bacillus pumilus gene specifying chloramphenicol acetyltransferase. Gene. 1983 Oct;24(2-3):163–169. doi: 10.1016/0378-1119(83)90076-8. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keggins K. M., Duvall E. J., Lovett P. S. Recombination between compatible plasmids containing homologous segments requires the Bacillus subtilis recE gene product. J Bacteriol. 1978 May;134(2):514–520. doi: 10.1128/jb.134.2.514-520.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsuk S., Ambulos N. P., Jr, Lovett P. S. Chloramphenicol-inducible gene expression in Bacillus subtilis is independent of the chloramphenicol acetyltransferase structural gene and its promoter. J Bacteriol. 1984 Oct;160(1):1–8. doi: 10.1128/jb.160.1.1-8.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsuk S., Chiang Y. W., Reynolds R. B., Lovett P. S. Restriction fragments that exert promoter activity during postexponential growth of Bacillus subtilis. J Bacteriol. 1983 Sep;155(3):1399–1406. doi: 10.1128/jb.155.3.1399-1406.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsuk S., Lovett P. S. Selective expression of a plasmid cat gene at a late stage of Bacillus subtilis sporulation. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3457–3460. doi: 10.1073/pnas.81.11.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Nicholson W. L., Chambliss G. H., Buckbinder L., Ambulos N. P., Jr, Lovett P. S. Isolation and expression of a constitutive variant of the chloramphenicol-inducible plasmid gene cat-86 under control of the Bacillus subtilis 168 amylase promoter. Gene. 1985;35(1-2):113–120. doi: 10.1016/0378-1119(85)90163-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoner R. G., Williams D. M., Lovett P. S. Enhanced expression of mouse dihydrofolate reductase in Bacillus subtilis. Gene. 1983 Apr;22(1):47–57. doi: 10.1016/0378-1119(83)90063-x. [DOI] [PubMed] [Google Scholar]
- Shaw W. V., Brenner D. G., LeGrice S. F., Skinner S. E., Hawkins A. R. Chloramphenicol acetyltransferase gene of staphylococcal plasmid pC221. Nucleotide sequence analysis and expression studies. FEBS Lett. 1985 Jan 1;179(1):101–106. doi: 10.1016/0014-5793(85)80200-3. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase: enzymology and molecular biology. CRC Crit Rev Biochem. 1983;14(1):1–46. doi: 10.3109/10409238309102789. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Yansura D. G., Henner D. J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci U S A. 1984 Jan;81(2):439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]




