Abstract
Folding mechanisms of proteins incorporated within supramolecular assemblies, including viruses, are little understood and may differ fundamentally from folding mechanisms of small globular proteins. We describe a novel Raman dynamic probe of hydrogen-isotope exchange to investigate directly these protein folding/assembly pathways. The method is applied to subunit folding in assembly intermediates of the double-stranded DNA bacteriophage P22. The icosahedral procapsid-to-capsid maturation (shell expansion) of P22 is shown to be accompanied by a large increase in exchange protection of peptide β-strands. The molecular mechanism of shell expansion involves unfolding of metastable tertiary structure to form more stable quaternary contacts and is governed by a surprisingly high activation energy. The results demonstrate that coat subunit folding and capsid expansion are strongly coupled processes. Subunit structure in the procapsid represents a late intermediate along the folding/assembly pathway to the mature capsid. Coupling of folding and assembly is proposed as a general pathway for the construction of supramolecular complexes.
Keywords: virus assembly/protein folding/H/D exchange/P22 phage/Raman spectroscopy
The assembly of a macromolecular complex from multiple copies of protein subunits requires intersubunit interactions that are highly specific. The strength of protein interactions, which determines the stability of the assembly, is roughly proportional to the sequestered surface area (1). Accordingly, the driving force for assembly is the sequestration of hydrophobic residues at subunit interfaces (2). Conversely, exposure of hydrophobic surfaces may compete with productive folding of subunits and lead to off-pathway aggregation rather than the native supramolecular architecture. A plausible mechanism for avoiding unproductive aggregation is assembly in several discrete steps with significant changes in subunit structure at each stage of the process and attainment of the native fold only within the mature assembly. In effect, some assembly intermediates may consist of subunits in incompletely folded states. These states are likely to possess compact shapes with well-defined secondary and tertiary structures, resembling the late-folding intermediates observed for small globular proteins (2). Such folding intermediates can be distinguished by the extent of protection of their peptide NH groups against deuterium exchange (3).
Until recently, the measurement of protium/deuterium (H/D) exchanges in proteins has been limited to relatively small globular structures that are amenable to study by high-field NMR spectroscopy. Current developments in time-resolved Raman spectroscopy offer a new approach for monitoring H/D exchanges in larger protein molecules and their complexes (4–8), thus facilitating the detection and characterization of protein-folding intermediates in supramolecular assemblies. Changes in secondary structure involving as few as 2% of residues can be measured accurately and interpreted structurally by use of Raman amide I and amide III bands, as exemplified recently for purified P22 coat (9) and scaffolding (10) molecules. In the present study, we have used the Raman dynamic probe to reveal differences in the fractions of deuterated peptide groups in assembly intermediates of the Salmonella bacteriophage P22, an icosahedral double-stranded DNA virus (11). Corresponding changes in subunit secondary structure along the pathway of P22 assembly are also characterized.
The assembly of phage P22 constitutes a model, multistep pathway for icosahedral viral morphogenesis (11, 12). In the initial step, 420 copies of the major coat protein (gp5, 429 residues, 47 kDa) assemble into a precursor procapsid in the presence of scaffolding protein subunits (gp8, 303 residues, 33 kDa). The metastable procapsid (diameter 58 nm) is subsequently transformed into the mature capsid (diameter 63 nm) concurrent with scaffolding release and DNA packaging, a process common to many other double-stranded DNA viruses, including the herpes viruses (13, 14). The P22 capsid transformation involves extensive rearrangements in the surface lattice and a 10% increase in radius (shell expansion) (15). However, this shell maturation is accompanied by only a small change in subunit secondary structure, despite large changes in side chain environments (9). These observations suggest that domain movement involving hinge bending mediates shell expansion (9, 15). Mild denaturants may also be used to extract scaffolding protein from procapsids without altering the coat protein lattice (15–17). Subsequent expansion of the empty procapsid shell can be induced in vitro either by heating to 65°C or by mild detergent treatment (17, 18).
The experiments reported here extend previous spectroscopic, calorimetric, and electron cryomicroscopic investigations of the P22 shell transformation and provide new insights into the molecular mechanisms of coat subunit folding and capsid maturation.
MATERIALS AND METHODS
Procapsids were prepared using Salmonella typhimurium strain DB7136 infected with P22 strain 2-am H200/13-am H101 as described (16) and further purified by Sephacryl S-1000 chromatography. Empty procapsid shells were prepared by repeated scaffolding (gp8) extraction with 0.5 M GuHCl at 4°C and sucrose gradient centrifugation.
Expanded shells were prepared from procapsid shells by heat treatment at 65°C (17). The progress of expansion was monitored by migration of particles on 1.2% agarose gels. The same protocol was used for shells assembled in D2O. The composition and integrity of particles were examined by SDS/PAGE and 1.2% Sea-Kem agarose gels.
Exchange experiments at 35°C were performed on samples twice pelleted and resuspended in isotopically pure D2O buffer. Samples were sealed in glass capillaries and allowed to complete fast exchange at 10°C. During subsequent incubation at 35°C, samples were periodically examined by Raman spectroscopy. At the end of each experiment (average duration, 3 wk), samples were examined for structural integrity (agarose gels) and composition (SDS/PAGE). Reversibility of local unfolding was confirmed by Raman spectroscopy on control samples that were incubated in H2O.
Subunits were refolded in D2O buffer as described by Teschke and King (19). Refolded deuterated subunits were assembled in the presence of scaffolding protein and purified by sucrose gradient centrifugation (16). Empty procapsid shells were prepared as described above.
Spectral measurements were performed on a Spex 1877 triple spectrograph equipped with a charge-coupled device array detector. The spectra were excited at 514.5 nm. Time-resolved spectra were collected at 1-min intervals. Time-resolved Raman spectra were normalized using the 1003 cm−1 band of phenylalanine as internal standard. Amide III′ bands were extracted by subtraction of the spectra of protiated particles. The resulting amide III′ difference bands were normalized with respect to the fully deuterated particles. The percentages of exchanged sites were calculated from these normalized amide III′ band areas. The protocol is fully described in previous papers (4–7).
The initial rates of procapsid shell expansion were obtained from separation of heat-treated products on 1.2% agarose gel (separation buffer: 40 mM Tris, 10 mM CH3COONa, 2 mM EDTA, pH 8.0) followed by Coomassie staining and densitometry.
RESULTS
Three Regimes for Exchange of Subunit Peptide (NH) Groups.
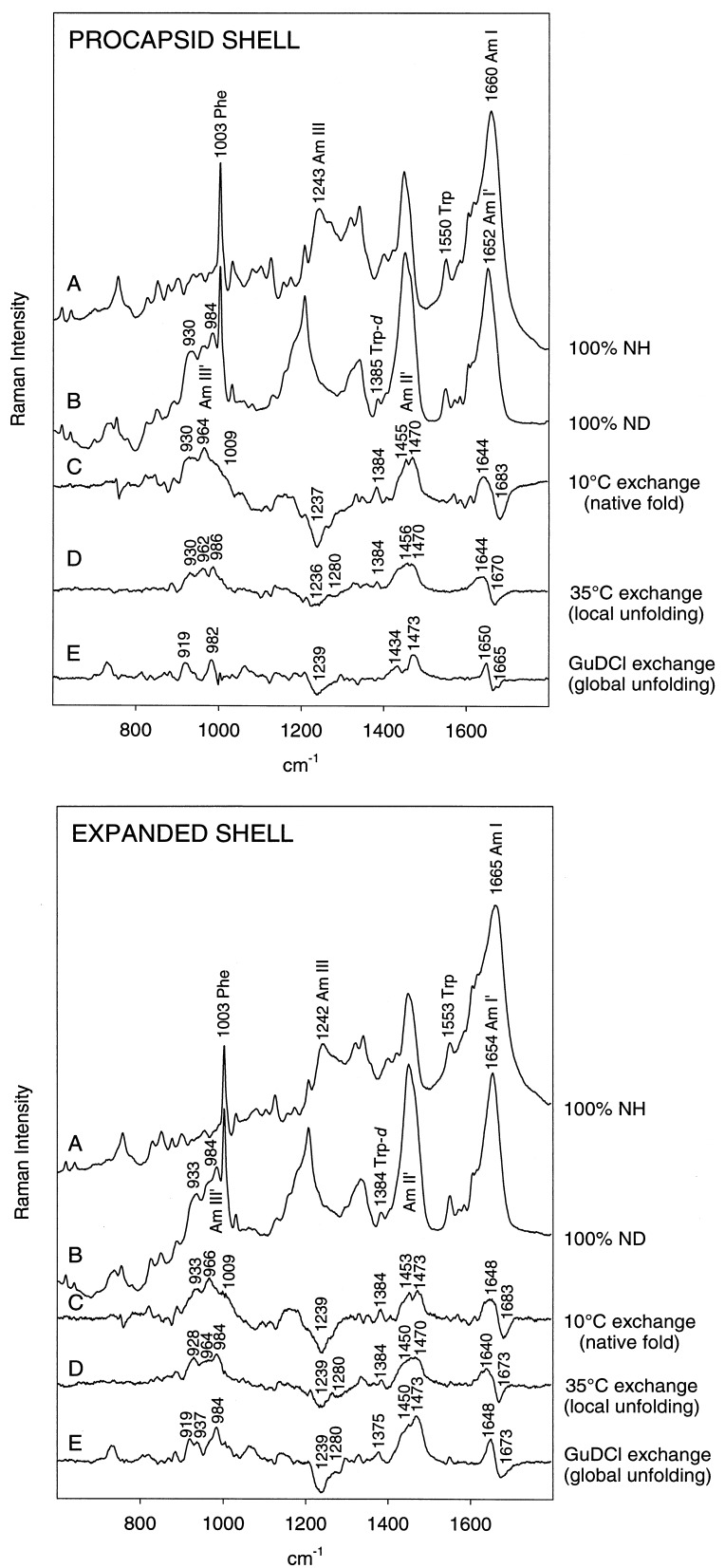
The effects of H/D exchange of peptide NH groups on Raman spectra of procapsid shells and expanded shells are illustrated in the Upper and Lower panels, respectively, of Fig. 1. For each type of shell, the Raman amide I band is centered near 1660–1665 cm−1 and the companion amide III band is centered near 1243 cm−1 (trace A in each panel of Fig. 1). The corresponding Raman amide bands for deuterated shells, designated amide I′ and amide III′, occur at approximately 1652 and 984 cm−1, respectively (trace B in each panel of Fig. 1). The positions of the amide I, amide I′, amide III, and amide III′ bands all indicate that β-strand is the dominant secondary structure of the coat protein subunit in both the procapsid shell and expanded shell assemblies, in accord with previous studies (9, 10).
Figure 1.
Raman monitoring of deuteration of peptide NH groups of procapsid (Upper) and expanded shells (Lower). In each panel: (A) H2O solution spectrum. (B) D2O solution spectrum of fully deuterated particle, obtained by refolding and reassembly in native D2O buffer of a sample previously treated with 6 M GuDCl. (C) Difference spectrum between A and particles exposed to D2O solution for 3 h at 10°C (signature of peptides exchanging from the native state). (D) Difference spectrum between particles exposed to D2O for 600 h at 35°C and particles exposed to D2O for 3 h at 10°C (signature of peptides exchanging via local unfolding). (E) Difference spectrum between B and particles exposed to D2O for 600 h at 35°C (signature of peptides in the exchange-protected core). Spectra were obtained from samples at 70 mg/ml concentration in 10 mM Tris buffer (pH 7.4) and 10°C.
We have identified three classes of exchangeable (NH → ND) peptide groups in shell subunits by varying the temperature or solvent environment at which Raman measurements are made. First, unprotected sites of shells maintained in native buffer at 10°C exchange relatively rapidly (19). These generate the Raman difference profile indicated as trace C in each panel of Fig. 1. A second class of sites, which is resistant to exchange at 10°C, can be exchanged in native buffer at 35°C via local unfolding (3). Heating to 35°C promotes partial unfolding of shell subunits without compromising the integrity of the shell assembly itself (17). This type of peptide exchange generates the Raman difference profile indicated as trace D in each panel of Fig. 1. Finally, a third class of peptide sites is resistant to exchange, even in shells containing partially unfolded (35°C) subunits. These are peptides of the exchange-protected core of the shell subunit. Amide protons of these peptides can be deuterated only by inducing shell disassembly and coat subunit unfolding in 6 M GuDCl buffer. Subsequent subunit refolding and shell reassembly in native D2O buffer permit structural characterization of the exchange-protected core by Raman difference spectroscopy, as shown in trace E of each panel of Fig. 1. We next discuss these three classes of peptide NH → ND exchange in turn.
Facile Exchange of Native Structure.
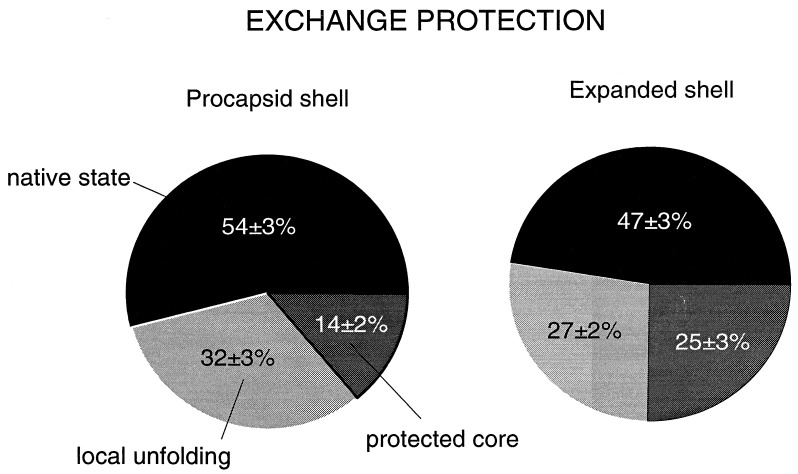
Relatively rapid exchange of peptide NH groups at 10°C is limited to domains of the shell subunits that are readily accessible to solvent and probably located on the surface of the assembly. With exchange of these residues, broad Raman bands appear at 930, 964, and 1009 cm−1, which are assignable to amide III′ markers of exposed peptide groups in loops, turns, and other irregular conformations (21). This conclusion is supported by the decrease of amide I intensity at 1683 cm−1 and concomitant increase of amide I′ intensity at 1640–1650 cm−1. Facile NH → ND exchange also occurs in α-helices and is manifested by Raman amide II′ intensity appearing near 1455 cm−1 in difference trace C. The amide II′ intensity adds to the prominent bands of the 1440–1460 cm−1 interval that are due to C-H bending modes of aliphatic side chains. All exchanges from the native state are complete within 3 h of exposure to D2O, both for procapsid shells and expanded shells. Facile NH → ND exchanges, including solvent-accessible peptides in loop, turn, and irregular conformations as well as rapidly exchanging peptides in native α-helical conformations, encompass 47 ± 3% of peptide sites in the expanded shell and 54 ± 3% of peptide sites of the procapsid shell (Fig. 2). The lower percentage of facile exchange in the expanded shell represents a small but significant increase in exchange protection upon shell expansion.
Figure 2.
Percentages of peptide NH groups exchanging in native, locally unfolded, and globally unfolded states of procapsid shells (54%, 32%, and 14%, respectively) and expanded shells (47%, 27%, and 25%, respectively). Exchange fractions were calculated from amide III′ band areas in the spectral interval 900-1020 cm−1. Exchange by local unfolding is defined as exchange that occurs at 35°C but not at 10°C. Exchange by global unfolding, which requires shell disassembly and subunit denaturation in 6 M GuDCl, defines peptides of the exchange-protected core.
Exchange of Labile Structure by Local Unfolding.
The Raman amide III′ signature of peptides exchanging via local unfolding at 35°C consists of two bands which are centered at 930 and 986 cm−1 in the procapsid shell (trace D, Fig. 1 Upper) and at 928 and 984 cm−1 in the expanded shell (trace D, Fig. 1 Lower). Intensities of these markers correlate with the intensity decreases of amide III in the interval 1230–1240 cm−1 and amide I at 1665–1675 cm−1, both of which are characteristic of β-strand (9, 10, 22). Similar correlation has been observed for the all β-strand coat protein P3 of bacteriophage PRD1. Thus, we assign amide III′ at 929/985 ± 1 cm−1 to peptides in the β-strand conformation. An additional amide III′ feature at 962 cm−1 in the procapsid shell spectrum indicates exchange from peptides with irregular secondary structure. The 962 cm−1 contribution is diminished in the expanded shell spectrum, indicating that the irregular structure is either protected or already exchanged at 10°C in this assembly. The latter interpretation is consistent with the exchange-protected core comprising exclusively β-strand (see next section). In both procapsid shells and expanded shells, the exchange of peptides via local unfolding is complete within 250 h of exposure to D2O. In the procapsid shell, 32 ± 3% of peptides exchange via local unfolding, whereas in the expanded shell only 27 ± 2% of sites are exchanged by this mechanism (Fig. 2).
Exchange of the Protected Core by Global Unfolding.
The Raman profile of the exchange-protected core is indicated for subunits of the procapsid shell and expanded shell in trace E of the appropriate panel of Fig. 2. In both types of shell, the Raman signature of the protected core consists of amide III′ bands near 919 and 982–984 cm−1, demonstrating that the exchange-protected core consists exclusively of β-strand. However, the amide III′ intensities are markedly different in the two types of shell, revealing a striking increase in the percentage of protected β-strand NH groups accompanying expansion. This interpretation is supported by the intensities observed for other deuteration-shifted amide bands. Thus, the shifted amide I → amide I′ and amide III → amide III′ bands are considerably more intense for the expanded shell than for the procapsid shell. On the basis of the measured amide intensities in traces E of Fig. 1, we have determined that the effective exchange-protected core of the procapsid shell subunit comprises 14 ± 2% of peptide groups, whereas that of the expanded shell subunit comprises 25 ± 3% of peptide groups (Fig. 2). Thus, the apparent exchange-protected core of the shell subunit nearly doubles with expansion.
Exchanges of Subunit Side Chains.
Tryptophans. Expansion of the P22 shell is accompanied by significant changes in environments and orientations of subunit side chains (9). The Raman spectrum is particularly sensitive to changes involving tryptophan, including differences in lability of the indole N1-H bond in procapsid shells and expanded shells. Indole N1H → N1D exchange, which serves as a gauge of tryptophan accessibility to solvent and local unfolding dynamics, is conveniently monitored by the intensity increase in the 1375–1385 cm−1 region of the Raman spectrum (23, 24). As seen in Fig. 1, the six tryptophans of the procapsid shell are more susceptible to native state exchange of N1H than are those of the expanded shell. We estimate that four to five Trp residues per subunit undergo facile (10°C) exchange, and another one to two exchange via local unfolding (35°C) in the procapsid shell. None of the six tryptophans of the procapsid shell is completely protected from exchange, i.e., there is no difference band near 1375–1385 cm−1 in trace E of the Upper panel of Fig. 1. Conversely, at least one and possibly two Trp residues resist exchange in the expanded shell, evidenced by the 1375 cm−1 difference band in trace E of the Lower panel of Fig. 1. Thus, one or two Trp side chains become part of the exchange-protected core upon shell expansion. This is consistent with previous data indicating a change from hydrophilic to hydrophobic local environment for the average Trp side chain following shell expansion (9).
Cysteine-405.
The single cysteine residue (Cys 405) of the P22 capsid subunit resists chemical modification of its sulfhydryl group in all assemblies investigated, including the coat protein monomer, procapsid shell, and expanded shell (P.E.P., unpublished results). Further information about the Cys 405 sulfhydryl environment can be obtained from the well-resolved and relatively sharp Raman band near 2570 cm−1, which is due to the S-H bond stretching vibration and is diagnostic of SH hydrogen bonding (25). The SH Raman band serves as a unique probe of local structure and dynamics (26). Fig. 3 shows that the band center differs significantly between procapsid and expanded shell forms, undergoing a shift from 2569 to 2573 cm−1 with expansion. The higher frequency and broader band shape associated with the expanded shell are indicative of weaker hydrogen bonding and a less hydrophobic sulfhydryl environment in the expanded shell than in the procapsid shell (25). In accord with this finding, we also observe more rapid SH → SD exchange of Cys 405 in the expanded shell than in the procapsid shell.
Figure 3.
Raman markers of the sulfhydryl group of Cys 405 in subunits of the procapsid shell (PS) and expanded shell (ES). (Right) The Raman S-H marker bands observed for H2O solutions of shells. (Left) The corresponding S-D markers observed for D2O solutions following complete SH → SD exchange.
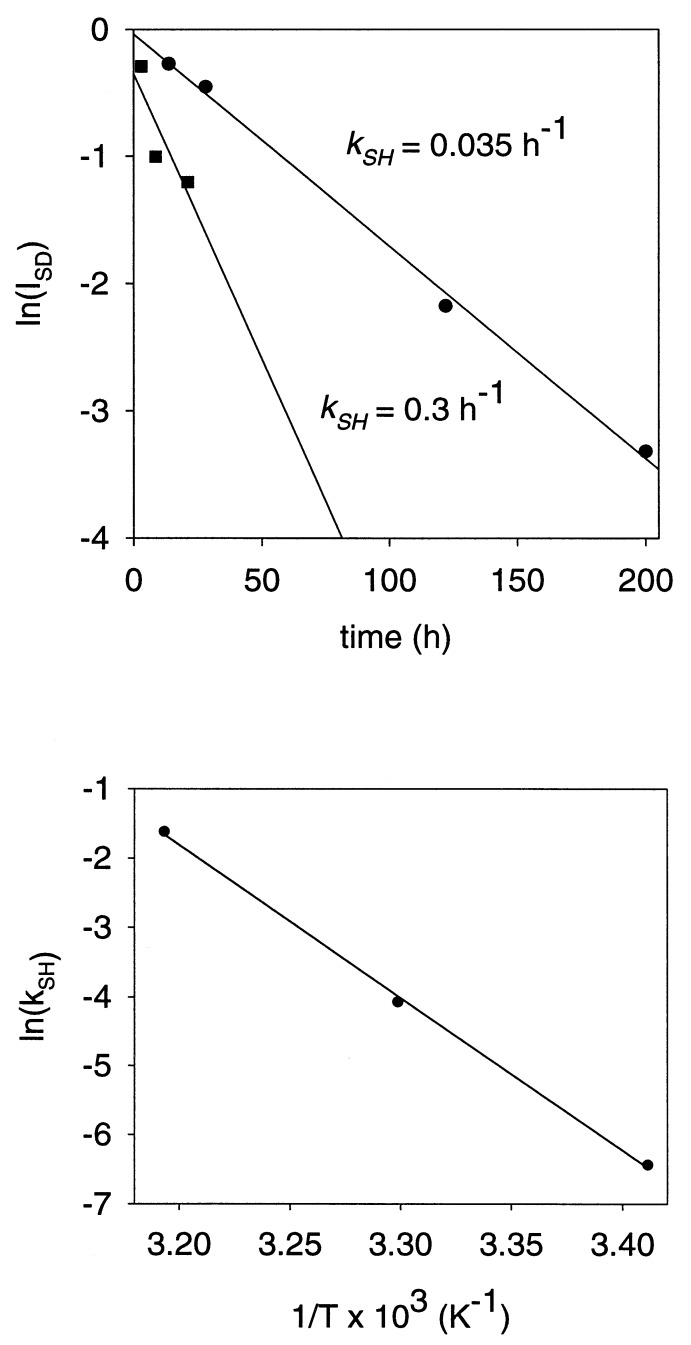
Although SH groups are well protected against exchange in the native procapsid (t < 20°C), SH → SD exchange is observed at a slightly elevated temperature, via local unfolding. For example, in the procapsid at 30°C, the sulfhydryl exchange rate constant (kSH) is measured as 0.035 h−1 (Fig. 4 Upper). The activation energy of local unfolding (Ealocal) calculated from the temperature dependence of procapsid SH exchange over the range 20–40°C is 44 kcal⋅mol−1 (1 kcal⋅mol−1 = 4.184 kJ⋅mol−1) (Fig. 4 Lower).
Figure 4.
(Upper) Rates (kSH) of Cys 405 sulfhydryl exchange in subunits of procapsid shells at 30°C (•) and expanded shells at 2°C (■). Data were obtained by measuring the intensity decay of the Raman S-H band of the shell as a function of time of exposure to D2O. (Lower) Arrhenius plot showing temperature dependence of the sulfhydryl exchange rate (kSH) in subunits of the procapsid shell. The slope of the plot corresponds to an activation energy for local unfolding (Ealocal) of 44 kcal⋅mol−1.
Conversely, for the expanded shell, SH → SD exchange is relatively facile at all temperatures. Thus, even at 2°C, we find for the expanded shell that kSH ≥ 0.3 h−1 (Fig. 4 Upper) and Ealocal is negligible in comparison to that of the procapsid. Interestingly, the Cys 405 sulfhydryl is less protected against exchange than is the average Trp indolyl group of the expanded shell.
Activation Energy of Shell Expansion.
To estimate the energy barrier opposing heat-induced shell expansion (Eaexpan), we measured the temperature dependence of the expansion rate constant, kexpan (Fig. 5 Upper). The Arrhenius plot of the measured rate constants (Fig. 5 Lower) exhibits considerable curvature, indicating significant temperature dependence of the apparent Eaexpan. We find Eaexpan = 40 ± 10 kcal⋅mol−1 in the range 40–55°C and Eaexpan = 90 ± 20 kcal⋅mol−1 in the range 55–70°C. Thus, the apparent activation energy of shell expansion at physiological temperature (37°C) is estimated as roughly equal to that of local unfolding, the latter determined from SH exchange dynamics (above). It should be noted that the expansion activation energy, Eaexpan, is expressed per mole of shells (420 subunits), while the local unfolding activation energy, Ealocal, is expressed per mole of subunits.
Figure 5.
(Upper) Kinetics of heat-induced procapsid expansion. Procapsid shells were heated at the indicated temperatures and aliquots of the reaction mixtures were withdrawn and separated on a 1.2% (wt/vol) agarose gel. Initial expansion rates (kexpan) were estimated from gel band intensities. (Lower) Arrhenius plot showing temperature dependence of the expansion rate.
DISCUSSION
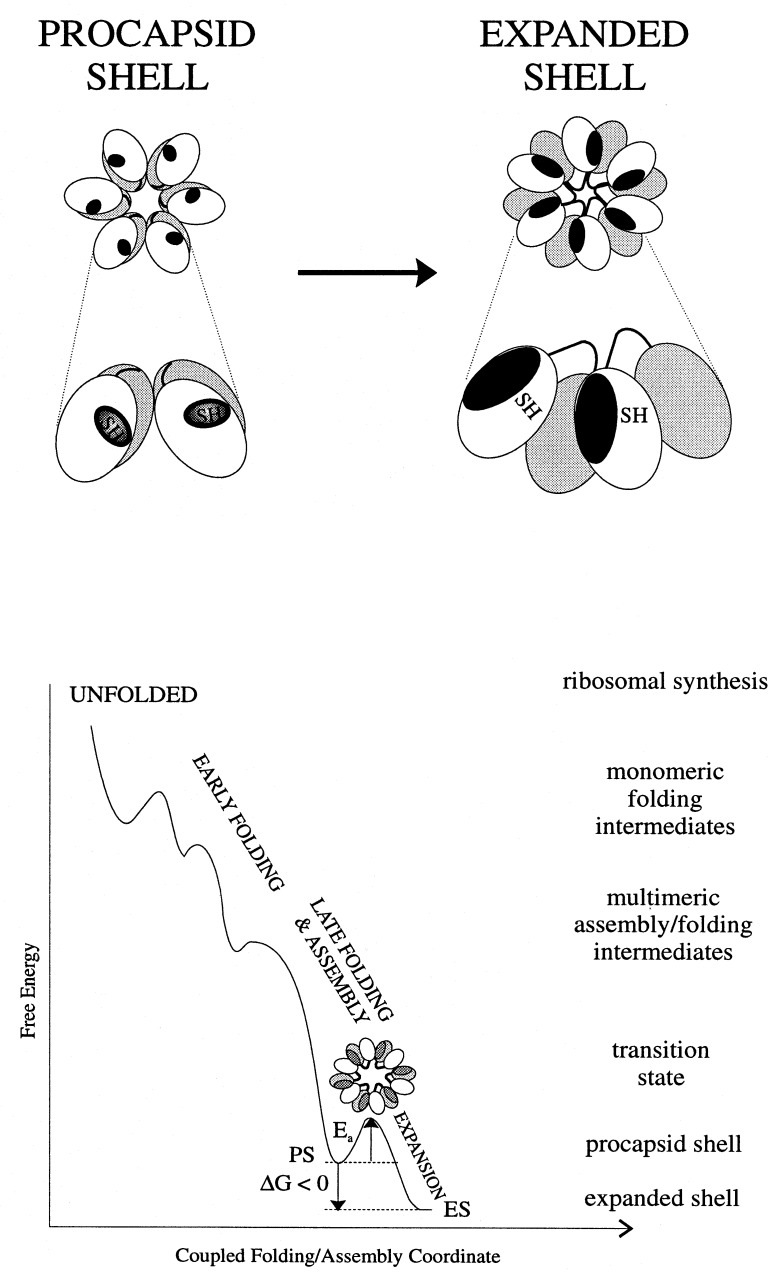
The striking increase in exchange protection that is observed with shell expansion (14% vs. 25%, Fig. 2) implies a corresponding increase in compactness of the icosahedrally arranged coat protein subunits. Similarly large increases in exchange protection are conferred by close packing of hydrophobic side chains during folding of the cores of typical globular proteins (27). Subunits of the mature (expanded) P22 shell may be viewed as representing the final native state of the coat protein, while those of the precursor shell are analogous to a late-folding intermediate. The native coat protein conformation, i.e., the state of lowest free energy, is thus attained only within the context of the expanded shell. An energy level diagram depicting coupling of coat protein folding with shell maturation is given in Fig. 6.
Figure 6.
(Upper) A mechanism accounting for increased exchange protection in the expanded shell (right) vis-à-vis the procapsid shell (left). The effective increase in the subunit exchange-protected core with expansion is due to domain interchange between neighboring subunits. The shell lattice (upper diagram) is represented as a cluster of six subunits (hexon) in which the protected core is indicated by dark shading and the two coat protein domains are shown as unshaded and lightly shaded. The enlargement (lower diagram) depicts rearrangement of contacts and domains, including Cys 405 sulfhydryls, between two neighboring subunits. (Lower) Energy landscape representation of coupling between subunit folding and capsid assembly in P22. A transition state containing partially exposed hydrophobic surfaces (dark gray) is proposed for the heat-induced expansion.
The present results could be explained by a simple protection mechanism in which hydrophobic residues that are exposed in the procapsid shell become sequestered in the expanded shell. However, such sequestration seems unlikely because the procapsid shell lacks exposed hydrophobic surfaces, as judged by its resistance to the reagents ANS and bisANS (28). A scheme that involves movement of coat protein domains, as illustrated in the upper panel of Fig. 6, is deemed more likely. Here, hydrophobic residues that form an intrasubunit interface between two coat protein domains in the procapsid shell become immobilized in the expanded shell. Evidence for domain structure of the coat protein has been obtained from unfolding studies (29). Immobilization is a consequence of domain interchange between neighboring subunits, wherein metastable tertiary contacts of procapsid subunits are replaced by more stable quaternary contacts of the expanded shell subunits. Such a model is supported by the Raman difference spectrum between procapsid and expanded shells, which reveals many changes in side chain environments despite a small change in subunit secondary structure (9). On the basis of the Raman signature observed for the protected core in the expanded shell subunit and in view of the relatively small change in secondary structure accompanying shell expansion, we conclude that the interchanged domain is mainly β-stranded. The domain interchange model proposed here is also supported by results of electron cryomicroscopy (15). The proposed mechanism of domain interchange during capsid expansion avoids excessive exposure of hydrophobic surfaces on isolated subunits, thus averting unproductive aggregation of subunits. At the same time, the stability of the mature assembly is not compromised.
The distinctive environments and different exchange dynamics observed for the Cys 405 sulfhydryl in procapsid and expanded shells suggest that Cys 405 is located at or near the interface between interchanging domains. If the activation energy of local unfolding (Ealocal = 44 kcal⋅mol−1, as measured by SH exchange) represents the free energy of domain opening, then simultaneous opening of the equivalent of one such interface would be required to account for the activation energy that triggers shell expansion at physiological temperature (Eaexp ≈ 35–40 kcal⋅mol−1 at 37°C). This explanation is based on the assumption that shell expansion is initiated at a single (but not necessarily unique) site in each shell and is propagated rapidly through the lattice. Such a mechanism is supported by the absence of partially expanded shells or other intermediates in both in vivo and in vitro preparations.
The apparent temperature dependence of the expansion activation energy (Fig. 5 Lower) may stem from temperature-dependent disruption of hydrophobic interactions. This would require transient exposure of hydrophobic residues to initiate expansion (Fig. 6), which is consistent with the previously reported acceleration of shell expansion by SDS (18). Alternatively, the expansion mechanism may involve a different transition state at higher temperatures.
The present results show conclusively that subunits in the procapsid shell of P22 are less protected against amide exchange than are those of the expanded shell. The subunit main chain in the procapsid shell presumably represents a more flexible conformation, which is appropriate to achieving the transition state. The driving force required to surmount the high activation energy barrier is evidently provided in vivo by ATP-driven DNA packaging. The in vitro effects of chemical agents such as SDS and urea may facilitate particular steps in the expansion, although at the present time the exact mechanism of their action is not known.
Finally, we note that formation of a metastable intermediate and subsequent transition to a stable final state has been observed for influenza virus hemagglutinin (30), in which the most stable state is attained only upon interaction with membrane at low pH. Interestingly, an irreversible, heat-induced, capsid expansion has been detected for poliovirus and other picornaviruses (31). This expansion is thought to correspond to capsid transformation during virus entry into the host cell (31). Since the most stable fold of subunits is attained only within the context of the mature assembly or during the membrane fusion, we propose that manipulation of the subunit folding pathway could provide a means for programming morphological transformations in such supramolecular ensembles.
Acknowledgments
Support from National Institutes of Health Grants GM50776 (to G.J.T.) and GM47980 (to P.E.P.) is gratefully acknowledged.
References
- 1.Janin J. Biochimie. 1995;77:497–505. doi: 10.1016/0300-9084(96)88166-1. [DOI] [PubMed] [Google Scholar]
- 2.Price N C. In: Mechanisms of Protein Folding. Pain R H, editor. New York: Oxford Univ. Press; 1994. pp. 160–193. [Google Scholar]
- 3.Bai Y, Sosnick T R, Mayne L, Englander S W. Science. 1995;269:192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li T, Johnson J E, Thomas G J., Jr Biophys J. 1993;65:1963–1972. doi: 10.1016/S0006-3495(93)81272-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reilly K E, Thomas G J., Jr J Mol Biol. 1994;241:68–82. doi: 10.1006/jmbi.1994.1474. [DOI] [PubMed] [Google Scholar]
- 6.Tuma R, Thomas G J., Jr Biophys J. 1996;71:3454–3466. doi: 10.1016/S0006-3495(96)79541-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuma R, Bamford J H K, Bamford D H, Russell M P, Thomas G J., Jr J Mol Biol. 1996;257:87–101. doi: 10.1006/jmbi.1996.0149. [DOI] [PubMed] [Google Scholar]
- 8.Tuma R, Thomas G J., Jr Biophys Chem. 1997;68:17–31. doi: 10.1016/s0301-4622(97)00001-x. [DOI] [PubMed] [Google Scholar]
- 9.Prevelige P E, Jr, Thomas D, Aubrey K L, Towse S A, Thomas G J., Jr Biochemistry. 1993;32:537–543. doi: 10.1021/bi00053a019. [DOI] [PubMed] [Google Scholar]
- 10.Tuma R, Prevelige P E, Jr, Thomas G J., Jr Biochemistry. 1996;35:4619–4627. doi: 10.1021/bi952793l. [DOI] [PubMed] [Google Scholar]
- 11.Prevelige P E, Jr, King J. Prog Med Virol. 1993;40:206–221. [PubMed] [Google Scholar]
- 12.King J, Chiu W. In: Structural Biology of Viruses. Chiu W, Burnett R M, Garcea R, editors. New York: Oxford Univ. Press; 1997. pp. 1–30. [Google Scholar]
- 13.Newcomb W W, Homa F L, Thomsen D R, Booy F P, Trus B L, Steven A C, Spencer J V, Brown J C. J Mol Biol. 1996;263:432–446. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- 14.Trus B L, Booy F P, Newcomb W W, Brown J C, Homa F L, Thomsen D R, Steven A C. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 15.Prasad B V V, Prevelige P E, Marietta E, Chen R O, Thomas D, King J, Chiu W. J Mol Biol. 1993;231:65–74. doi: 10.1006/jmbi.1993.1257. [DOI] [PubMed] [Google Scholar]
- 16.Prevelige P E, Jr, Thomas D, King J. J Mol Biol. 1988;202:743–757. doi: 10.1016/0022-2836(88)90555-4. [DOI] [PubMed] [Google Scholar]
- 17.Galisteo M L, King J. Biophys J. 1993;65:227–235. doi: 10.1016/S0006-3495(93)81073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Earnshaw W, Casjens S, Harrison S C. J Mol Biol. 1976;104:387–410. doi: 10.1016/0022-2836(76)90278-3. [DOI] [PubMed] [Google Scholar]
- 19.Teschke C M, King J. Biochemistry. 1993;32:10839–10847. doi: 10.1021/bi00091a040. [DOI] [PubMed] [Google Scholar]
- 20.Finucane M D, Jardetzky O. Protein Sci. 1996;5:653–662. doi: 10.1002/pro.5560050409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugawara J, Harada I, Matsuura H, Shimanouchi T. Biopolymers. 1978;17:1405–1421. [Google Scholar]
- 22.Chen M C, Lord R C. J Am Chem Soc. 1974;96:4750–4752. doi: 10.1021/ja00822a004. [DOI] [PubMed] [Google Scholar]
- 23.Miura T, Takeuchi H, Harada I. Biochemistry. 1988;27:88–94. doi: 10.1021/bi00401a015. [DOI] [PubMed] [Google Scholar]
- 24.Miura T, Takeuchi H, Harada I. J Raman Spectrosc. 1989;20:667–671. [Google Scholar]
- 25.Li H, Thomas G J., Jr J Am Chem Soc. 1991;113:456–462. [Google Scholar]
- 26.Li H, Hanson C, Fuchs J A, Woodward C, Thomas G J., Jr Biochemistry. 1993;32:5800–5808. doi: 10.1021/bi00073a012. [DOI] [PubMed] [Google Scholar]
- 27.Kamtekar S, Schiffer J M, Xiong H, Babik J M, Hecht M H. Science. 1993;262:1680–1685. doi: 10.1126/science.8259512. [DOI] [PubMed] [Google Scholar]
- 28.Teschke C M, King J, Prevelige P E., Jr Biochemistry. 1993;32:10658–10665. doi: 10.1021/bi00091a016. [DOI] [PubMed] [Google Scholar]
- 29.Prevelige P E, Jr, King J, Silva J L. Biophys J. 1994;66:1631–1641. doi: 10.1016/S0006-3495(94)80955-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carr C M, Chaudhry C, Kim P S. Proc Natl Acad Sci USA. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chow M, Basavappa R, Hogle J M. In: Structural Biology of Viruses. Chiu W, Burnett R M, Garcea R, editors. New York: Oxford Univ. Press; 1997. pp. 1–30. [Google Scholar]