Abstract
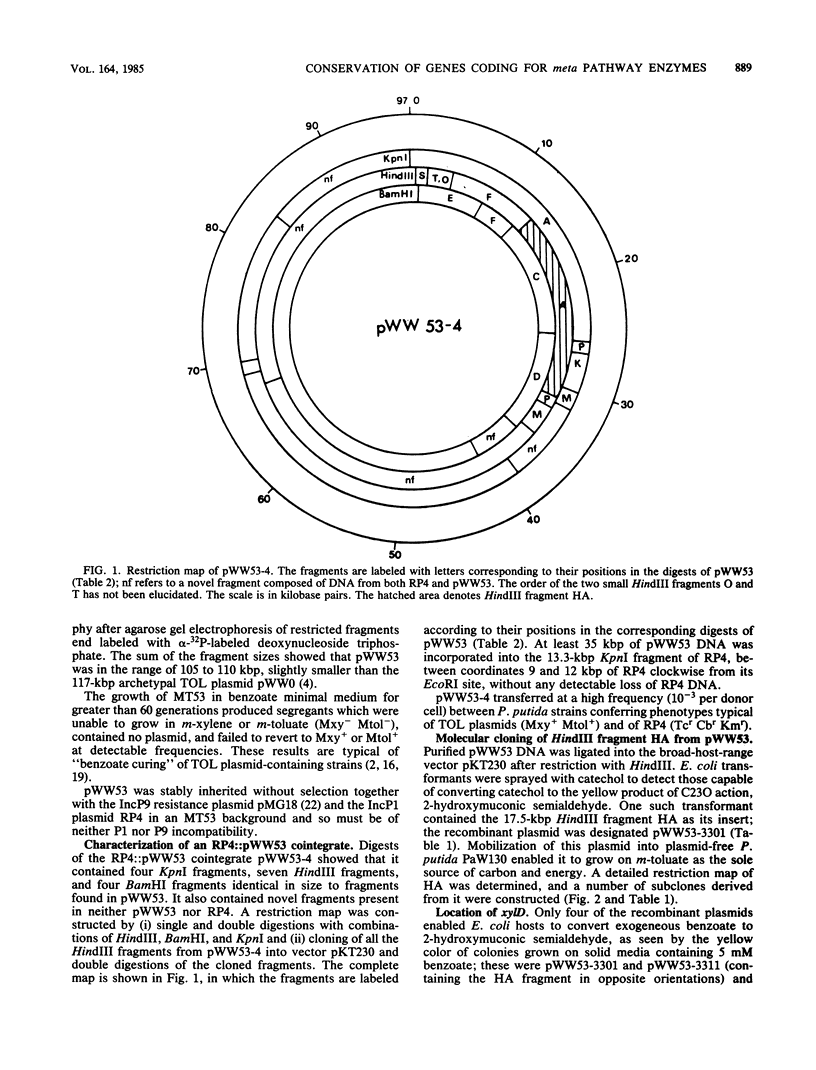
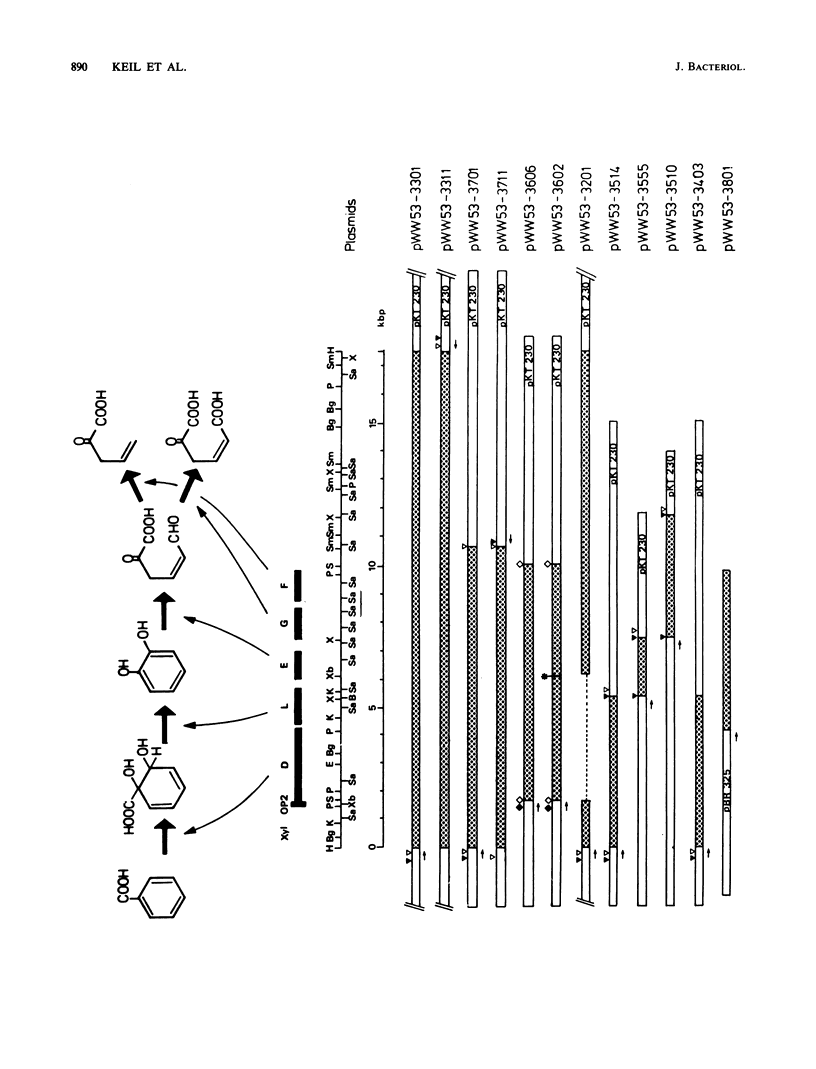
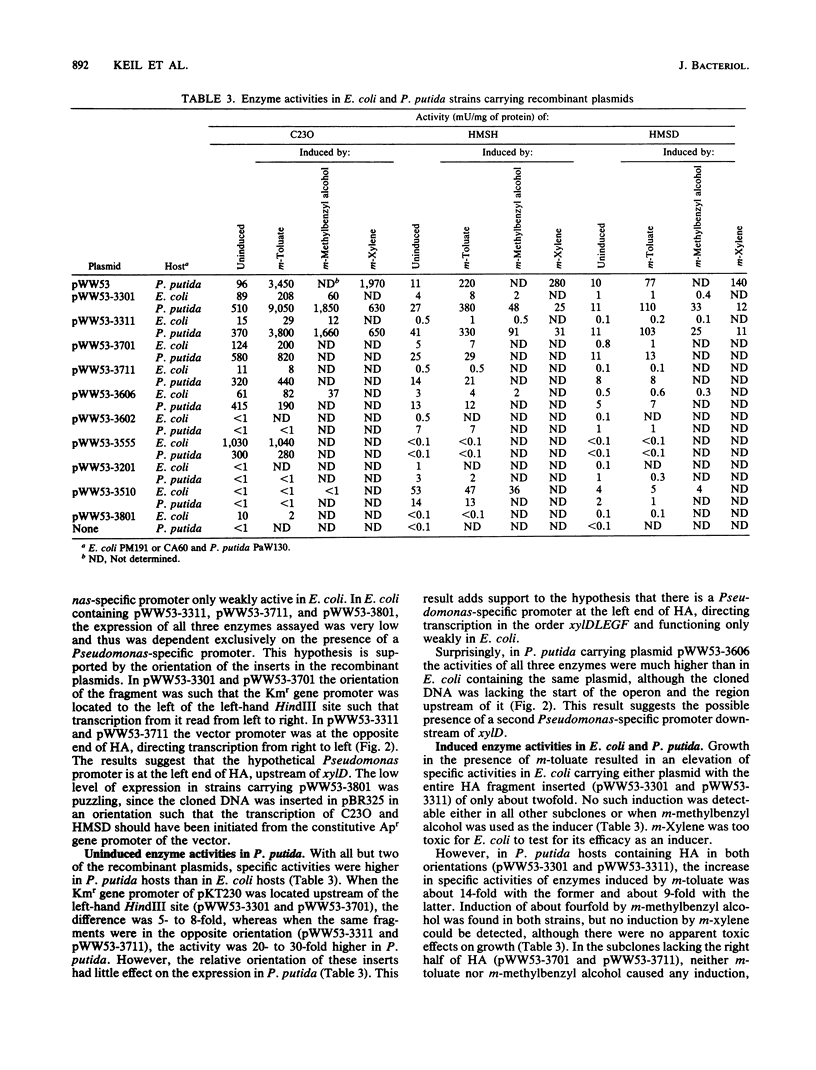
Pseudomonas putida MT53 contains a TOL plasmid, pWW53, that encodes toluene-xylene catabolism. pWW53 is nonconjugative, is about 105 to 110 kilobase pairs (kbp) in size, and differs significantly in its restriction endonuclease digestion pattern and incompatibility group from the archetypal TOL plasmid pWW0. An RP4::pWW53 cointegrate plasmid, pWW53-4, containing about 35 kbp of pWW53 DNA, including the entire catabolic pathway genes, was formed, and a restriction map for KpnI, HindIII, and BamHI was derived. The entire regulated meta pathway genes for the catabolism of m-toluate were cloned into pKT230 from pWW53 on a 17.5-kbp HindIII fragment. The recombinant plasmid supported growth on m-toluate when mobilized into plasmid-free P. putida PaW130. A restriction map of the insert for 10 restriction enzymes was derived, and the locations of xylD, xylL, xylE, xylG, and xylF were determined by subcloning and assaying for their gene products in both Escherichia coli and P. putida hosts. Good induction of the enzymes by m-toluate and m-methylbenzyl alcohol but not by m-xylene was measured in P. putida, but little or no regulation was found in E. coli. The restriction map and the gene order showed strong similarities with published maps of the DNA encoding both the entire meta pathway operon (xylDLEGFJIH) and the regulatory genes xylS and xylR on the archetype TOL plasmid pWW0, suggesting a high degree of conservation in DNA structure for the catabolic operon on the two different plasmids.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., Timmis K. N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981 Dec;16(1-3):237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Bayley S. A., Duggleby C. J., Worsey M. J., Williams P. A., Hardy K. G., Broda P. Two modes of loss of the Tol function from Pseudomonas putida mt-2. Mol Gen Genet. 1977 Jul 20;154(2):203–204. doi: 10.1007/BF00330838. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing R., Broda P. A cleavage map of the TOL plasmid of Pseudomonas putida mt-2. Mol Gen Genet. 1979;177(1):189–191. doi: 10.1007/BF00267270. [DOI] [PubMed] [Google Scholar]
- Duggleby C. J., Bayley S. A., Worsey M. J., Williams P. A., Broda P. Molecular sizes and relationships of TOL plasmids in Pseudomonas. J Bacteriol. 1977 Jun;130(3):1274–1280. doi: 10.1128/jb.130.3.1274-1280.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Bagdasarian M., Bagdasarian M. M., Timmis K. N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Lehrbach P. R., Lurz R., Rueckert B., Bagdasarian M., Timmis K. N. Localization and functional analysis of transposon mutations in regulatory genes of the TOL catabolic pathway. J Bacteriol. 1983 May;154(2):676–685. doi: 10.1128/jb.154.2.676-685.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinter N. J. A broad-host-range cloning vector transposable to various replicons. Gene. 1983 Jan-Feb;21(1-2):133–143. doi: 10.1016/0378-1119(83)90155-5. [DOI] [PubMed] [Google Scholar]
- Harayama S., Lehrbach P. R., Timmis K. N. Transposon mutagenesis analysis of meta-cleavage pathway operon genes of the TOL plasmid of Pseudomonas putida mt-2. J Bacteriol. 1984 Oct;160(1):251–255. doi: 10.1128/jb.160.1.251-255.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Molecular cloning of gene xylS of the TOL plasmid: evidence for positive regulation of the xylDEGF operon by xylS. J Bacteriol. 1981 Nov;148(2):413–418. doi: 10.1128/jb.148.2.413-418.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Molecular cloning of regulatory gene xylR and operator-promoter regions of the xylABC and xylDEGF operons of the TOL plasmid. J Bacteriol. 1983 Sep;155(3):1192–1199. doi: 10.1128/jb.155.3.1192-1199.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Nucleotide sequence of the promoter region of the xylDEGF operon on TOL plasmid of Pseudomonas putida. Gene. 1984 Sep;29(3):323–330. doi: 10.1016/0378-1119(84)90061-1. [DOI] [PubMed] [Google Scholar]
- Jeenes D. J., Williams P. A. Excision and integration of degradative pathway genes from TOL plasmid pWW0. J Bacteriol. 1982 Apr;150(1):188–194. doi: 10.1128/jb.150.1.188-194.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil H., Lebens M. R., Williams P. A. TOL plasmid pWW15 contains two nonhomologous, independently regulated catechol 2,3-oxygenase genes. J Bacteriol. 1985 Jul;163(1):248–255. doi: 10.1128/jb.163.1.248-255.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz D. A., Chapman P. J. Isolation and characterization of spontaneously occurring TOL plasmid mutants of Pseudomonas putida HS1. J Bacteriol. 1981 Jun;146(3):952–964. doi: 10.1128/jb.146.3.952-964.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickup R. W., Williams P. A. Spontaneous deletions in the TOL plasmid pWW20 which give rise to the B3 regulatory mutants of Pseudomonas putida MT20. J Gen Microbiol. 1982 Jul;128(7):1385–1390. doi: 10.1099/00221287-128-7-1385. [DOI] [PubMed] [Google Scholar]
- Prentki P., Krisch H. M. A modified pBR322 vector with improved properties for the cloning, recovery, and sequencing of blunt-ended DNA fragments. Gene. 1982 Feb;17(2):189–196. doi: 10.1016/0378-1119(82)90072-5. [DOI] [PubMed] [Google Scholar]
- Sala-Trepat J. M., Evans W. C. The meta cleavage of catechol by Azotobacter species. 4-Oxalocrotonate pathway. Eur J Biochem. 1971 Jun 11;20(3):400–413. doi: 10.1111/j.1432-1033.1971.tb01406.x. [DOI] [PubMed] [Google Scholar]
- Summers A. O., Jacoby G. A. Plasmid-determined resistance to boron and chromium compounds in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1978 Apr;13(4):637–640. doi: 10.1128/aac.13.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatcroft R., Williams P. A. Rapid methods for the study of both stable and unstable plasmids in Pseudomonas. J Gen Microbiol. 1981 Jun;124(2):433–437. doi: 10.1099/00221287-124-2-433. [DOI] [PubMed] [Google Scholar]
- Williams P. A., Murray K. Metabolism of benzoate and the methylbenzoates by Pseudomonas putida (arvilla) mt-2: evidence for the existence of a TOL plasmid. J Bacteriol. 1974 Oct;120(1):416–423. doi: 10.1128/jb.120.1.416-423.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P. A., Worsey M. J. Ubiquity of plasmids in coding for toluene and xylene metabolism in soil bacteria: evidence for the existence of new TOL plasmids. J Bacteriol. 1976 Mar;125(3):818–828. doi: 10.1128/jb.125.3.818-828.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Franklin F. C., Williams P. A. Regulation of the degradative pathway enzymes coded for by the TOL plasmid (pWWO) from Pseudomonas putida mt-2. J Bacteriol. 1978 Jun;134(3):757–764. doi: 10.1128/jb.134.3.757-764.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Metabolism of toluene and xylenes by Pseudomonas (putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975 Oct;124(1):7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K., Nishi T. pKJ1, a naturally occurring conjugative plasmid coding for toluene degradation and resistance to streptomycin and sulfonamides. J Bacteriol. 1980 Aug;143(2):552–560. doi: 10.1128/jb.143.2.552-560.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]



