Abstract
Targeting of protein cargo to the vacuole/lysosome is a multistep process that appears to have conserved features between mammalian, yeast, and plant cells. In each case, some soluble vacuolar/lysosomal proteins are believed to be bound by transmembrane cargo receptors in the trans-Golgi network (TGN) that redirect these proteins into clathrin-coated vesicles. These vesicles then appear to be transported to the prevacuole/endosome by a trafficking machinery that requires components identified in other vesicle-targeting steps such as N-ethylmaleimide-sensitive factor (NSF), soluble NSF attachment protein (SNAP), SNAP receptors (SNAREs), rab-type GTPases, and Sec1p homologs. Two likely members of this trafficking machinery have been characterized from Arabidopsis thaliana: AtPEP12p, a t-SNARE that resides on a what we now call a prevacuolar compartment, and AtELP, a protein that shares many common features with mammalian and yeast transmembrane cargo receptors. Here, we have further investigated the intracellular distribution of AtELP. We have found that AtELP is located at the trans-Golgi of Arabidopsis root cells, and that its C terminus can preferentially interact in vitro with the mammalian TGN-specific AP-1 clathrin–adapter complex, suggesting a likely role in clathrin-coated, vesicle-directed trafficking at the TGN. Further, consistent with a role in trafficking of vacuolar cargo, we have found that AtELP partially colocalizes with AtPEP12p on a prevacuolar compartment.
Most vacuolar/lysosomal proteins first enter the secretory pathway at the endoplasmic reticulum (ER), then travel through the Golgi apparatus to the trans-Golgi network (TGN). At the TGN, vacuolar proteins are sorted away from secreted proteins because of the presence of positive vacuolar sorting signals (VSS). These VSS are posttranslationally added mannose-6-phosphate residues in mammalian cells, or specific protein sequences in plant and yeast cells (reviewed in refs. 1 and 2). Plants appear to have a much more complex set of independent VSS, and the plant VSS do not function in yeast cells (reviewed in ref. 1). This specificity may be a result of the requirement for specific cargo receptors that recognize each VSS. These cargo receptors have been characterized from several eukaryotic cells and include the mammalian mannose-6-phosphate receptor (3), the yeast carboxypeptidase Y (CPY) receptor (Vps10p; ref. 4), and the putative plant vacuolar sorting receptor (BP-80; ref. 5). Though overall sequence homology between these receptors is low, all are type-I membrane proteins that share common structural features such as luminal cysteine-rich repeats, a single transmembrane spanning domain near the C terminus, and the presence of Tyr-based motifs (Yxxφ, where Y stands for Tyr; x, for any residue; and φ, for a bulky, hydrophobic residue) on the cytoplasmic tail. In mammalian cells, soluble cargo bound for the lysosome is packaged into clathrin-coated vesicles (CCVs; ref. 6), and this is believed to be the case for soluble vacuolar hydrolases in yeast, as well (7). In plant cells, it has been reported that some vacuolar-targeted proteins leave the TGN in CCVs (8, 9); however, trafficking of proteins to the plant vacuole is a complex process, and other vacuolar proteins have been shown to depart the TGN in vesicles that are different from CCVs (9). Proteins destined for secretion have not been found to leave the TGN in CCVs (2, 6).
CCVs are formed at both the plasma membrane (PM) and the TGN. Research in mammalian cells has indicated that these can be differentiated because of specific clathrin adaptor complexes (APs) found in each. APs connect the clathrin coat to the vesicle membrane through interaction with the Tyr motifs (or other signals) present in the cytoplasmic tail of receptors and other membrane proteins. Endocytosed proteins and receptors are packaged at the PM by AP-2, whereas proteins at the TGN are packaged by AP-1 (reviewed in ref. 10). Homologs of the mammalian AP complexes have been characterized in yeast (11–13), and potential homologs of some proteins in the AP complex have been identified in plants (14–16), suggesting that the adaptor specificity seen in mammalian cells may be conserved. After budding, the coat disassembles and vesicles are apparently trafficked to the prevacuolar compartment (PVC) using machinery similar to that used in other vesicular trafficking steps such as N-ethylmaleimide-sensitive factor (NSF), soluble NSF attachment protein (SNAP), SNAP receptors (SNAREs), rab-type GTPases, and Sec1p homologs (the SNARE hypothesis, reviewed in refs. 17 and 18). This has been shown best in yeast for the transport of CPY by Vps10p: vesicles that carry CPY are thought to contain the vesicle-(v-)SNARE Vti1p (19), which directs delivery to the PVC through interaction with the PVC target-(t-)SNARE Pep12p as well as several soluble factors including NSF and the Sec1p-homolog Vps45p (19, 20). CPY then continues on to the vacuole through a process that requires the vacuolar t-SNARE Vam3p (21), whereas Vps10p (22, 23) and presumably Vti1p (19) are recycled back to the Golgi apparatus.
Although the plant vacuolar targeting pathway appears to be much more complex than that found in yeast cells, both in the diversity of VSS as well as the type and number of vacuole-bound vesicles that leave the TGN, some conserved features will be found in all eukaryotic cells. Consistent with this hypothesis, several homologs of the machinery described above for yeast cells have been found in plants. AtPEP12p, an Arabidopsis thaliana homolog of yeast Pep12p, has been cloned and found to reside on a PVC in root cells (24, 25). AtVAM3p, an Arabidopsis protein that complements a deletion mutant of the yeast vacuolar t-SNARE Vam3p, has been identified (26). AtVPS45p, a Sec1p homolog from Arabidopsis that can functionally replace Vps45p in yeast has been characterized recently (27). Further, a putative cargo receptor isolated from pea (Pisium sativum) CCVs that has been shown to bind the vacuolar sorting signals of some proteins (BP-80; ref. 5) also has been described. We have previously isolated AtELP, a protein from Arabidopsis, which shares all the features common to cargo receptors found throughout eukaryotes (cysteine-rich repeats, single transmembrane domain, and cytoplasmic Tyr-based motifs) and is associated with CCVs isolated from peas (28). AtELP is highly homologous to BP-80, suggesting that it also may play a role in targeting of proteins to the plant vacuole, as has been suggested for BP-80 (5, 29). Here, we have investigated further the association of AtELP with CCVs and have found that its cytoplasmic tail is capable of in vitro interaction with the proteins of the mammalian TGN-specific AP-1 adaptor complex. Further, through both biochemical approaches and immunoelectron microscopy, we have found that AtELP is localized on the trans-Golgi, as well as being colocalized with the t-SNARE AtPEP12p on the PVC of Arabidopsis root cells.
MATERIALS AND METHODS
In Vitro AP-1 Adaptor Complex-Binding Assay.
AP-1 and AP-2 adaptor complexes were purified from calf brain-coated vesicles as described previously (30, 31). Details of the synthesis of the biotinylated photoactivatable cross-linking *Lamp1-YQTI and *TGN38-YQRL peptides are described elsewhere (30, 31). Peptides corresponding to the cytoplasmic tail of AtELP (YMPL) and the Tyr-606 → Ala mutated form (AMPL) represent residues 599–612 within the AtELP sequence (accession no. U86700) and were synthesized at the Macromolecular Structure Facility at Michigan State University by using an Applied Biosystems 432A peptide synthesizer. The peptides were purified by gel filtration on a Biogel P2 column and analyzed by mass spectrometry and microsequencing. UV-induced cross-linking of the photoreactive peptides to purified AP complexes in the absence or presence of varying amounts of the AtELP competitor peptides was carried out as described (30). After the cross-linking reaction, the cross-linked products were treated with sample buffer under reducing and denaturing conditions, analyzed by SDS/PAGE, and transferred to nitrocellulose. The membranes were probed with streptavidin-horse radish peroxidase and detected by enhanced chemiluminescence.
Sucrose Density Gradients.
Arabidopsis thaliana plants, ecotype RLD, were grown in liquid culture as described in ref. 25. Roots (≈2 g) were separated from leaves and stems, chilled to 4°C, and ground in 6 ml of 50 mM Hepes⋅KOH, pH 6.5/5 mM EDTA/13.7% (wt/vol) sucrose/0.1 mM phenylmethylsulfonyl fluoride/1 mM DTT. The extracts were cleared at 2,000 × g before layering of 3 ml on top of a preformed step gradient consisting of the following steps: 1.0 ml 54%, 2.7 ml 40%, 2.2 ml 33%, 2.0 ml 24%, and 1.5 ml 15% (wt/vol) sucrose in 50 mM Hepes⋅KOH, pH 6.5/5 mM EDTA. The gradients were spun in a Beckman SW40 swinging bucket rotor at 150,000 × g for 3 hr at 4°C. Twenty-four 0.5-ml fractions were collected from the top, and sucrose concentrations for each fraction were determined with a refractometer. All fractions were then precipitated with 10% trichloroacetic acid, solubilized in SDS sample buffer, and equal volumes of each fraction were analyzed by SDS/PAGE. After transfer to nitrocellulose, proteins were detected with specific antisera to AtELP (28), AtPEP12p (25), the tonoplast marker pyrophosphatase (32), and Golgi marker ARA-4p (33). For quantification of results, blots were digitized on a flat-bed scanner and densitometry was accomplished by using nih image software. The resulting data for each marker were normalized to their respective backgrounds, and the percentage of marker protein detected in each fraction with respect to the total loaded in all fractions was calculated.
Electron Microscopy.
Cryosections of Arabidopsis roots were done essentially as described previously (25). For immunolabeling, grids were floated on drops in successive solutions at room temperature according to Slot et al. (34). After blocking in 1% BSA in PBS, sections were incubated with primary antibodies to AtPEP12p or AtELP for 4 hr. Excess antibody was removed by multiple washes of 1% BSA in PBS. Primary antibodies were detected by biotinylated goat anti-rabbit IgG for 1 hr and then streptavidin-conjugated to 10-nm colloidal gold particles. For double-labeling, the grids first were treated as above for AtPEP12p antibodies, then a second fixation step using 1% glutaraldehyde and 0.02% Gly followed by a second blocking step with 1% BSA in PBS was utilized to prevent cross-reactivity of the AtPEP12-antisera in later steps (34). The grids then were incubated with either no antisera or specific antisera for AtELP for 4 hr, followed by a 1-hr incubation with goat anti-rabbit IgG linked directly to 5-nm colloidal gold particles. The grids were washed in distilled water and stained according to Griffiths et al. (35). The sections were observed with a Hitachi model H600 electron microscope operating at 75 kHz. All labeling experiments were conducted several times each on independent sections. Quantification of immunogold staining was done by counting the number of gold particles found within a 0.33-μm2 circle centered on either the Golgi, the cytoplasm, or the nuclear matrix of many independent micrographs.
RESULTS
AtELP Is Localized to the Trans-Golgi and Post-Golgi Membranes.
The first step at which protein targeting to the vacuole/lysosome differs from secretion occurs in the TGN, where cargo receptors such as the mannose-6-phosphate receptor of mammalian cells and CPY receptor (Vps10p) of yeast bind to their specific cargo and redirect it away from the bulk flow of proteins in the secretory pathway (3, 4). Thus, the first step in investigating the role of AtELP as a potential cargo receptor was to examine cryosections of Arabidopsis roots for the localization of AtELP.
The majority of the AtELP-associated labeling was found on electron-dense, uncoated vesicular structures that often were found near the trans-Golgi of the root cells (Fig. 1A, arrowheads). These structures were similar to the post-Golgi structure identified previously for AtPEP12p in Arabidopsis roots (ref. 25; see below). Significant AtELP labeling was also found to be associated with the Golgi apparatus (Fig. 1B, small arrows). Statistical analysis of many independent micrographs indicated that the level of Golgi labeling was 9-fold higher than that of the cytoplasm or nuclei; further, almost all of the Golgi-associated labeling was found on the trans-side. The orientation of the Golgi was determined based on appearance and the more electron-dense staining pattern of the trans-Golgi. Some lower level of labeling also was found associated with more cis- or medial-stacks of the Golgi (data not shown), though this may represent AtELP in transit to the trans-Golgi. AtELP antisera was not found to label the ER, PM, or tonoplast (data not shown). Labeling with the preimmune sera was not seen on the Golgi or the post-Golgi structures and mainly consisted of random background staining (Fig. 1C). The fixation and cryosectioning conditions used here are designed to maintain antigenicity of membrane proteins and are not optimal for preservation of coated vesicles; thus, we were unable to determine directly whether AtELP was associated with CCVs as has been shown biochemically (28). These results showed that AtELP is localized to the trans-face of the Golgi in Arabidopsis root cells, as well as being found in other structures that are likely post-Golgi membranes.
Figure 1.
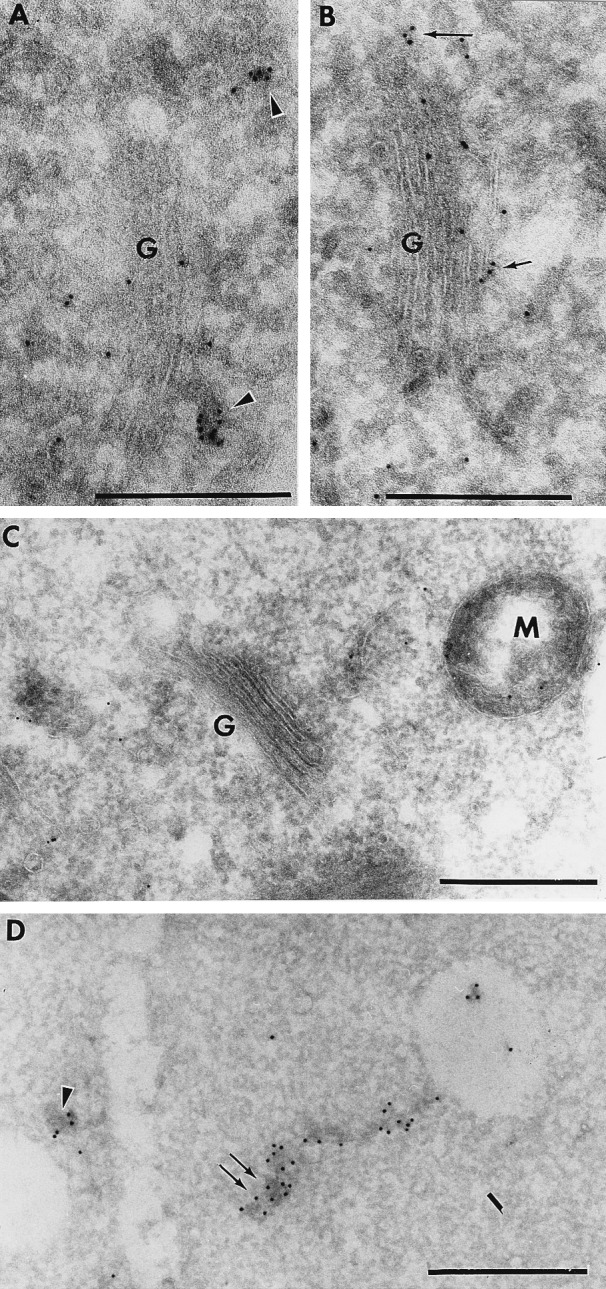
AtELP is localized on the trans-Golgi and post-Golgi membranes in cryosections of Arabidopsis roots. Sections were treated with antisera to AtELP (A and B), preimmune for AtELP (C), or with antisera to AtPEP12p (D) followed by biotinylated goat anti-rabbit secondary antibodies. Antibodies were then detected by streptavidin conjugated to 10 nm colloidal gold. G, Golgi; M, mitochondria. (Bar = 0.5 μm.)
We have further investigated the localization of AtPEP12p and have found that, in addition to the 100-nm electron-dense uncoated vesicles reported earlier (ref. 25; Fig. 1D, arrowhead), AtPEP12p labeling also was associated with electron-dense reticulotubular compartments with diameters of approximately 100 nm (Fig. 1D, double arrow). These tubules were found to label heavily with AtPEP12p antisera and occasionally were found to attach to small electron-lucent membranes that are perhaps small vacuoles. Because these tubules share the same electron-dense staining pattern and a similar diameter (100 nm), it is possible that the previously described vesicular structures (Fig. 1D, arrowhead) represent cross-sections through these tubules. We now refer to both structures as a PVC. No staining of the PVC was seen with preimmune sera (ref. 25 and data not shown).
The Cytoplasmic Tail of AtELP Interacts Specifically with the TGN-Associated AP-1 Adaptor Complex.
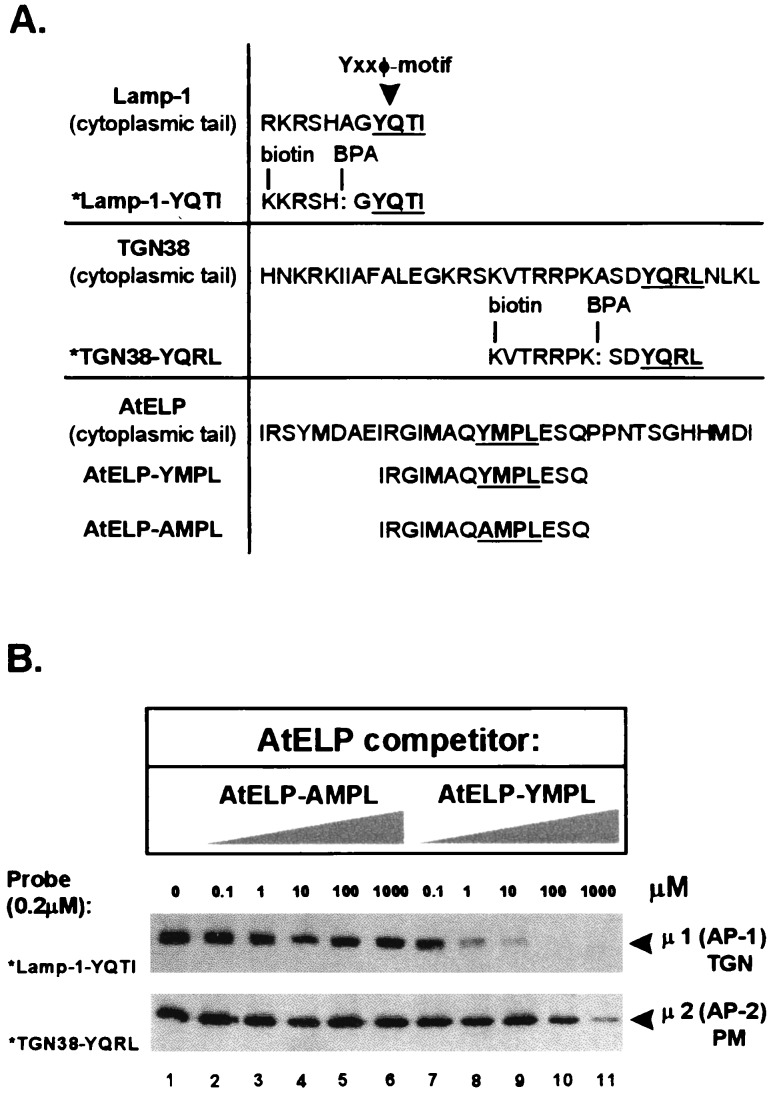
Considering that AtELP has been reported to be associated with CCVs (28) and that its cytoplasmic tail contains a potential Tyr-motif (YMPL, residues 606–609), it was therefore likely that AtELP would interact with the adaptor complex as part of forming CCVs. The recognition of a Tyr-motif by either the AP-1 or AP-2 complex depends on the sequence context surrounding the Tyr-motif (10, 31); thus, to determine which type of AP complex is capable of interacting with the cytoplasmic tail of AtELP, we performed an in vitro assay. Unfortunately, although some components of the plant APs have been identified by sequence homology (14–16), they have not been characterized biochemically. So, considering the likely conservation in the AP mechanisms, we chose to use the well defined mammalian AP complexes. The μ-subunits of both the TGN-associated AP-1 and the PM-associated AP-2 adaptor complex of mammalian cells have been shown to interact with the Tyr-motifs of several mammalian proteins such as Lamp-1 and TGN38 (30, 31, 36). Recent studies have shown the specificity of AP-1 for the Tyr-motif of Lamp-1 (31) and the specificity of AP-2 for TGN38 (30). In these assays, photoactivatable cross-linking is used to show binding of a biotinylated Lamp-1 or a TGN38 cytoplasmic-tail peptide to the μ-subunit of the AP complexes, which subsequently is detected with streptavidin conjugated to horseradish peroxidase. Using this assay, we addressed whether synthetic peptides corresponding to the cytoplasmic tail of AtELP would compete for (and therefore block) binding of the AP complexes to Lamp-1 or TGN38 peptides.
The synthetic peptides used in this study are shown in Fig. 2A. The biotinylated, photoactivatable *Lamp1-YQTI and *TGN38-YQRL tail peptides have been described previously (30, 31). The AtELP peptides correspond to residues 599–612 of the cytoplasmic tail (YMPL) or to this sequence with Tyr-606 changed to Ala (AMPL) to potentially inactivate the Tyr-motif. In the absence of AtELP peptides, 100% binding of the Lamp-1 and TGN38 peptides was seen to μ1 of the AP-1 and to μ2 of the AP-2 complexes, respectively (Fig. 2B, lane 1). The mutated AtELP-AMPL peptide was unable to compete for binding of either μ1 or μ2 (Fig. 2B, lanes 2–6) to *Lamp1-YQTI or *TGN38-YQRL (respectively). One micromolar concentration of the AtELP-YMPL peptide was sufficient for half-maximal competition, and 10 μM abolished greater than 90% of the binding of μ1 (AP-1) to the *Lamp1-YQTI peptide (Fig. 2B Middle, lanes 7–11), indicating that the cytoplasmic tail of AtELP likely was binding to the AP-1 complex with high affinity. Only at a 1,000-fold-higher concentration was the AtELP-YMPL peptide able to compete away binding of μ2 (AP-2) to the *TGN38-YQRL peptide (Fig. 2B Bottom, lanes 7–11). Hence, the AtELP peptide appears to have a higher affinity for μ1 of the TGN-specific AP-1 than for μ2 of the PM-specific AP-2 complex. Further, that the AtELP-AMPL peptide was unable to block binding suggests that the Tyr-motif of AtELP is required for interaction with the AP complex. These results, together with the localization of AtELP to the trans-Golgi of Arabidopsis roots (see above), suggests that AtELP may have a role in CCV-directed trafficking of proteins at the TGN of plant cells.
Figure 2.
Interaction of the cytoplasmic tail of AtELP with the AP-1 complex. A schematic representation of peptides used for the cross-linking studies involving AP-1 and AP-2 complexes is shown in A. Complete sequences of the Lamp-1, TGN38, and AtELP cytoplasmic tails containing the Yxxφ motif are compared with the sequences of the synthetic peptides used in this study. The Tyr-based motifs are highlighted and underlined. The *Lamp1-YQTI and *TGN38-YQRL peptides were modified by replacement of an alanine with the UV-photoactivatable, cross-linking agent benzoylphenylalanine (BPA), and by addition of biotin to the N terminus. AP-1 and AP-2 complexes (0.2 mg/ml) purified from calf brain-coated vesicles were incubated with 0.2 μM of either *Lamp1-YQTI (B Middle) or *TGN38-YQRL (B Bottom) cross-linking peptides and increasing amounts (0–1,000 μM) of the AtELP-YMPL or mutated AtELP-AMPL peptides as competitors. The appearance of the cross-linked photoreactive peptides to μ1 of AP-1 (Middle) or μ2 of AP-2 (Bottom) upon UV irradiation was tested after separation by SDS/PAGE using streptavidin conjugated to horseradish peroxidase.
AtPEP12p and AtELP Partially Cofractionate in Sucrose Density Gradients.
Given that AtPEP12p appears to be a t-SNARE residing on a PVC and that AtELP is a potential vacuolar cargo receptor, it seemed likely that these proteins would at least partially reside on the same membrane population. To determine whether this was the case, we subjected Arabidopsis root tissue to sucrose density gradients optimized to provide better separation of plant endomembranes (see Materials and Methods) and examined the fractionation pattern of AtPEP12p, AtELP, and several available markers for the plant endomembrane system, with antisera specific for each protein.
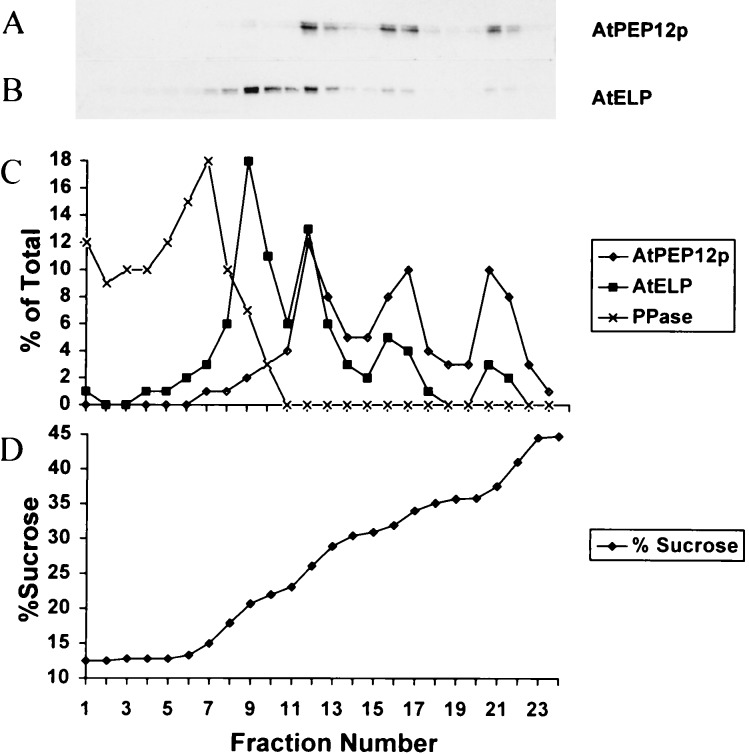
AtPEP12p can be differentiated into three distinct peaks at densities of approximately 1.12, 1.14, and 1.17 g/ml (Fig. 3A). AtELP was found to have a major low-density peak at 1.08 g/ml, with additional minor peaks found at higher densities of 1.12, 1.14, and 1.17 g/ml (Fig. 3B). Importantly, it appeared that AtPEP12p and AtELP cofractionated in the 1.12, 1.14, and 1.17 g/ml peaks, a result that was seen more clearly after quantification of the fractions by densitometry (Fig. 3C). Neither protein cofractionated with pyrophosphatase (Fig. 3C), a marker for the tonoplast (29). In addition, the fractionation pattern of AtPEP12p and AtELP was distinct from ARA-4p (data not shown), a marker for the Golgi (30). The peaks observed for these proteins are unlikely to represent the residual interfaces of the step gradient because (i) after centrifugation, the density profile of the gradient is virtually linear (Fig. 3D), and (ii) the densities of the peaks observed for these proteins are not consistent with the proteins accumulating at an interface between steps. Thus, the cofractionation of AtELP and AtPEP12p in the higher-density peaks suggests that these two proteins reside on the same compartment, which likely represents a PVC. Meanwhile, the low-density peak (1.08 g/ml), which only contained AtELP, may represent the trans-Golgi staining seen for AtELP (see Fig. 1B) or potentially may be a population of vesicles in transit to or from the PVC.
Figure 3.
Equilibrium density gradient analysis of Arabidopsis roots. Postnuclear membranes were loaded onto a sucrose step gradient and spun to equilibrium. Twenty-four fractions were collected from the top and trichloroacetic acid-precipitated, and equal volumes of each fraction were separated by SDS/PAGE and transferred to nitrocellulose. AtPEP12p (A), AtELP (B), and pyrophosphatase were detected by using specific antisera to each protein. Blots were analyzed by densitometry, and the percentage of the total marker protein detected in each fraction for AtPEP12p, AtELP, and pyrophosphatase was plotted in C. The sucrose concentration of each fraction was determined by refractometry and was plotted in D.
AtELP and AtPEP12p Colocalize on a PVC.
We have shown above that AtELP was localized to the trans-Golgi as well as to electron-dense post-Golgi membranes in cryosections of Arabidopsis roots. These electron-dense vesicles were similar to the late post-Golgi organelle (now called a PVC) that contained AtPEP12p (ref. 25; Fig. 1D, arrowhead). Together with the cofractionation of AtELP and AtPEP12p in density gradients, this suggested that AtELP and AtPEP12p likely would colocalize in these structures. To investigate this possibility, we performed double-labeling experiments on cryosections of Arabidopsis roots in which AtPEP12p was first labeled with specific antisera and detected with 10 nm gold. A second fixation and blocking step was then performed before incubating the sections with either no antisera or antisera specific to AtELP, followed by detection with 5 nm gold.
We found that both AtPEP12p and AtELP antisera specifically labeled many of the same electron-dense, uncoated vesicular structures (Fig. 4, arrowheads). No labeling of these structures with 5 nm gold was seen in the absence of AtELP antisera (data not shown). Fig. 4 represents a glancing section through the Golgi apparatus, and thus, it is difficult to clearly identify the trans-face. Still, some vesicles were found to only be labeled with AtELP (5 nm gold), and these appeared to be associated with the Golgi (Fig. 4, small arrow). These results indicated that some populations of AtELP coexists with AtPEP12p on the same membrane population, and the results are consistent with the biochemical cofractionation reported above. Further, considering the likely role of AtELP in trafficking of vacuolar proteins from the TGN, the colocalization of AtELP with AtPEP12p on the electron-dense structures supports our argument that these membranes represent a PVC.
Figure 4.
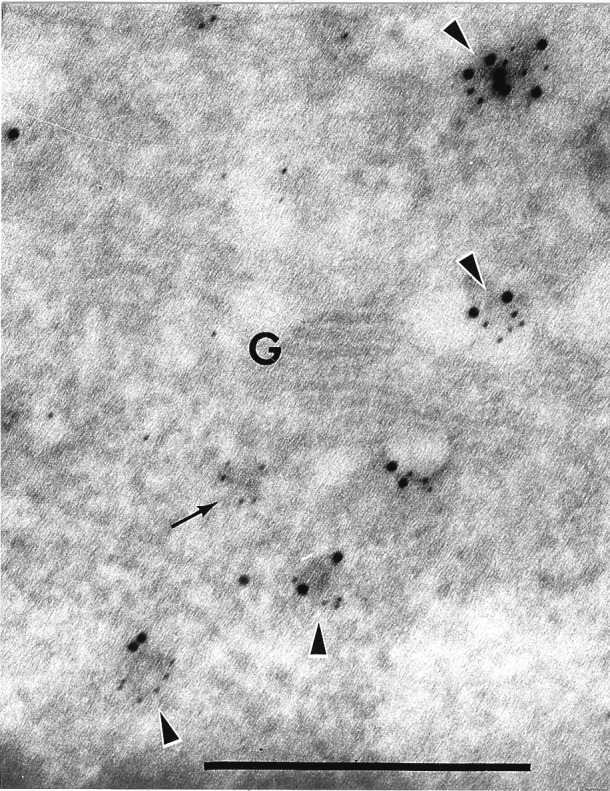
AtELP and AtPEP12p colocalize on a PVC in cryosections of Arabidopsis roots. Section was incubated with antisera to AtPEP12p, followed by biotinylated goat anti-rabbit secondary antibodies, then visualized with streptavidin conjugated to 10 nm colloidal gold. After a second fixation step (see Materials and Methods), the same sections were incubated with antibodies to AtELP, followed by goat anti-rabbit secondary antibodies conjugated to 5 nm colloidal gold. G, Golgi. (Bar = 0.5 μm.)
DISCUSSION
The plant vacuole is a vital, multifunctional organelle with roles in protein storage, osmoregulation, cell growth, and development (reviewed in ref. 2). Trafficking of proteins to the vacuole has been studied extensively, and the signals involved in targeting to the plant vacuole have been well defined (37). Some components of this trafficking machinery have begun to be characterized, including putative cargo receptors involved in vacuolar targeting (BP-80, ref. 29; AtELP, ref. 28), as well as proteins that are likely to function in vesicle fusion steps during trafficking to the vacuole: the t-SNAREs AtPEP12p (24, 25) and AtVAM3p (26), and the Sec1p-homologue AtVPS45p (27). Here, we have reported on further characterization of the plant vacuolar targeting machinery. We have shown that AtELP, consistent with a proposed role as a cargo receptor for vacuolar cargo proteins and its biochemical association with CCVs, is localized at the trans-Golgi and can interact in vitro with the proteins of the mammalian TGN-specific AP-1 adaptor complex. Further, we have shown through both biochemical and immunocytochemical means that a population of AtELP colocalizes with the t-SNARE AtPEP12p on a membrane compartment that is likely to be a PVC.
Our results have shown that the cytoplasmic Tyr-based motif of AtELP can compete efficiently for the mammalian AP-1 with the Tyr-based motif of a protein known to function at the TGN (Lamp-1). It is important to note that AtELP has much less affinity for the mammalian AP-2 complex, which argues against an involvement of AtELP in endocytic CCVs at the plasma membrane. Although a few components of the adaptor complexes have been identified in plants (14–16), they have not been well characterized. Moreover, it has not been possible to clearly distinguish whether these components are members of the AP-1 or AP-2 complexes of plant cells. Thus, it is not yet possible to carry out experiments similar to those shown here using the plant adaptor complexes. Regardless, the overall conservation apparent in the adaptor complexes among various cell types suggests that our results using the mammalian adaptor complex are most likely relevant to the function of AtELP in plants. The immunological localization of AtELP to the trans-Golgi, together with the fact that AtELP has been found associated with CCVs (28), suggest that AtELP may function in CCV-directed trafficking of vacuolar proteins at the TGN of plant cells.
The cofractionation of AtPEP12p and AtELP in sucrose density gradients is supported by the immunogold-electron microscopy. Based on the unique fractionation pattern of AtPEP12p in density gradients as well as immunoelectron microscopy (25), we have suggested previously that AtPEP12p represents a novel marker for a PVC. The fact that we now find AtELP, a protein possibly involved in transport of vacuolar cargo, associated with AtPEP12p further supports that this organelle represents a PVC in plant cells. It is interesting that the PVC that is described here for AtPEP12p has such a heterogeneous fractionation pattern in density gradients (i.e., three peaks). It is possible that the PVC is a fragile organelle that fragments during the isolation and centrifugation procedures, although the consistent fractionation pattern observed from many independent experiments (data not shown) makes this seem unlikely. The heterogeneity also could be explained by the apparent complexity of the vacuolar system in plant cells. At least four biochemically distinct pathways for targeting of soluble and membrane proteins to the plant vacuole have been described (reviewed in ref. 1). Moreover, some plant cells have been reported to have two types of vacuoles, which each receive different cargo proteins (9, 38). Thus, it is possible that AtPEP12p resides on many separable PVCs as part of its role in receiving protein traffic.
It is also intriguing to speculate that the low-density (1.08 g/ml) peak of AtELP, which does not appear to cofractionate with AtPEP12p, may represent a biochemical marker for the TGN in plant cells, a result that is supported by the electron microscopic studies that found only AtELP associated with the trans-Golgi whereas AtELP and AtPEP12p appear to consistently colocalize on PVCs. Although we find some AtELP labeling on the Golgi by electron microscopy, our results have shown that AtELP does not cofractionate with the Golgi marker ARA-4p; nor did AtELP cofractionate with the Golgi-associated enzymatic activity of fucosyl transferase (28). This may suggest that AtELP passes quickly through the Golgi and cannot be found there at steady state (i.e., in density gradients). Alternately, the trans-Golgi membranes stained by AtELP may represent a biochemically distinct membrane from the Golgi-proper. Or, it may also be that ARA-4p and fucosyl transferase are restricted to the cis-side of the Golgi in roots and, thus, sediment in the gradient as a distinct population of membranes. Clearly, a more thorough examination of the biochemical markers for the plant Golgi is required. It is also possible that this low-density peak may represent vesicles in transit to or from the PVC that contain AtELP. Further experiments are needed to clearly identify the nature of this vesicle population.
Our data argues for a role for AtELP in transporting cargo from the TGN to the PVC; yet, the nature of the cargo that AtELP may be carrying remains unclear. AtELP is clearly homologous to BP-80 (72% identical), a protein shown to associate with the N-terminal VSS of some soluble vacuolar-targeted proteins (5). However, an AtELP/BP-80 homolog recently has been characterized from pumpkin (PV72/82) that binds in vitro to both a C-terminal and internal, but not N-terminal, VSS from pumpkin pro2S albumin (39). We have not been able to show an in vitro association of AtELP with either the N-terminal VSS of sweet potato sporamin or the C-terminal VSS of barley lectin (S.U.A., M. Bar-Peled, and N.V.R., unpublished observations), though this perhaps is because of the lack of posttranslational modification and improper folding of the bacterially expressed protein used in these studies. Together with the fact that there are several AtELP-like genes found in the Arabidopsis EST database (28), as well as two full-length pea BP-80-like clones identified by Paris et al. (29), this suggests that AtELP, pea BP-80, and pumpkin PV72/82 may be members of a family of related vacuolar-sorting receptors in plant cells. That each may have different affinities for each type of VSS may provide the specificity required for the sorting of multiple proteins to the vacuole by different pathways.
In conclusion, we have examined further the components of the vacuolar-targeting machinery in plants. As outlined above, our findings suggest that AtELP is involved in CCV-directed trafficking of cargo proteins from the TGN of plant cells to the PVC defined by AtPEP12p. The exact nature of this prevacuolar organelle is still unclear because of the complexity of the vacuolar pathway in plant cells. However, the connection between these two markers, as well as to other endomembrane markers that we continue to characterize, will allow us to investigate this complexity in greater detail.
Acknowledgments
We acknowledge Diane Bassham and Maor Bar-Peled for valuable comments on the manuscript. A.A.S. is a National Institute of Health Postdoctoral Fellow (GM18861). N.V.R. is supported by the National Science Foundation (MCB-9507030) and the Department of Energy (DE-FG02-91ER-20021). T.K. is supported by the National Institutes of Health (GM36548) as well as by Special Funds from the Center for Blood Research at Harvard Medical School. I.R. is supported by Special Funds from the Department of Cell Biology at Harvard Medical School. F.M. and D.M.-M. are supported by Ministère de l’Enseignement Supérier et de la Recherche (DSPT5-EA 469), the Centre National de Recherche Scientifique (DSV27), the Consiel Régionel de Bourgogne, and the Délégation Régionele à la Recherche Scientifique et à la Technologie.
ABBREVIATIONS
- AP
clathrin adaptor complex
- CCV
clathrin coated vesicle
- ER
endoplasmic reticulum
- PM
plasma membrane
- PVC
prevacuolar compartment
- TGN
trans-Golgi network
- SNARE
soluble N-ethylmaleimide sensitive factor attachment protein receptor
- VSS
vacuolar sorting signal
References
- 1.Bassham D C, Raikhel N V. Adv Bot Res. 1997;25:43–58. [Google Scholar]
- 2.Robinson D G, Hinz G. Protoplasma. 1997;197:1–25. [Google Scholar]
- 3.Kornfeld S. Annu Rev Biochem. 1992;61:307–330. doi: 10.1146/annurev.bi.61.070192.001515. [DOI] [PubMed] [Google Scholar]
- 4.Marcusson E G, Hordazdovsky B F, Cereghino J L, Gharakhanian E, Emr S D. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- 5.Kirsch T, Paris N, Butler J M, Beevers L, Rogers J C. Proc Natl Acad Sci USA. 1994;91:3403–3407. doi: 10.1073/pnas.91.8.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Traub L M, Kornfeld S. Curr Opin Cell Biol. 1997;9:527–533. doi: 10.1016/s0955-0674(97)80029-4. [DOI] [PubMed] [Google Scholar]
- 7.Seeger M, Payne G S. EMBO J. 1992;11:2811–2818. doi: 10.1002/j.1460-2075.1992.tb05348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harley S M, Beevers L. Plant Physiol. 1989;91:674–678. doi: 10.1104/pp.91.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hohl I, Robinson D G, Chrispeels M J, Hinz G. J Cell Sci. 1996;109:2539–2550. doi: 10.1242/jcs.109.10.2539. [DOI] [PubMed] [Google Scholar]
- 10.Kirchhausen T, Bonifacino J S, Riezman H. Curr Opin Cell Biol. 1997;9:488–495. doi: 10.1016/s0955-0674(97)80024-5. [DOI] [PubMed] [Google Scholar]
- 11.Phan H L, Finlay J A, Chu D S, Tan P K, Kirchhausen T, Payne G S. EMBO J. 1994;13:1706–1717. doi: 10.1002/j.1460-2075.1994.tb06435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rad M R, Phan H L, Kirchrath L, Tan P K, Kirchhausen T, Hollenberg C P, Payne G S. J Cell Sci. 1995;108:1605–1615. doi: 10.1242/jcs.108.4.1605. [DOI] [PubMed] [Google Scholar]
- 13.Stepp J D, Pellicena-Palle A, Hamilton S, Kirchhausen T, Lemmon S K. Mol Biol Cell. 1995;6:41–58. doi: 10.1091/mbc.6.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holstein S E, Drucker M, Robinson D G. J Cell Sci. 1994;107:945–953. doi: 10.1242/jcs.107.4.945. [DOI] [PubMed] [Google Scholar]
- 15.Maldonado-Mendoza I E, Nessler C L. Plant Mol Biol. 1996;32:1149–1153. doi: 10.1007/BF00041398. [DOI] [PubMed] [Google Scholar]
- 16.Maldonado-Mendoza I E, Nessler C L. Plant Mol Biol. 1997;35:865–872. doi: 10.1023/a:1005923829224. [DOI] [PubMed] [Google Scholar]
- 17.Bennet M K. Curr Opin Cell Biol. 1995;7:581–586. doi: 10.1016/0955-0674(95)80016-6. [DOI] [PubMed] [Google Scholar]
- 18.Hay J C, Scheller R H. Curr Opin Cell Biol. 1997;9:505–512. doi: 10.1016/s0955-0674(97)80026-9. [DOI] [PubMed] [Google Scholar]
- 19.Fischer von Mollard G, Nothwehr S F, Stevens T H. J Cell Biol. 1997;137:1511–1524. doi: 10.1083/jcb.137.7.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burd C G, Peterson M, Cowles C R, Emr S D. Mol Biol Cell. 1997;8:1089–1104. doi: 10.1091/mbc.8.6.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darsow T, Rieder S E, Emr S D. J Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cereghino J L, Marcusson E G, Emr S D. Mol Biol Cell. 1995;6:1089–1102. doi: 10.1091/mbc.6.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper A A, Stevens T H. J Cell Biol. 1996;133:529–542. doi: 10.1083/jcb.133.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bassham D C, Gal S, Conceição A S, Raikhel N V. Proc Natl Acad Sci USA. 1995;92:7262–7266. doi: 10.1073/pnas.92.16.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conceição A S, Marty-Mazars D, Bassham D C, Sanderfoot A A, Marty F, Raikhel N V. Plant Cell. 1997;9:571–582. [PMC free article] [PubMed] [Google Scholar]
- 26.Sato M H, Nakamura N, Oshumi Y, Kouchi O, Kondo M, Hara-Nishimura I, Nishimura M, Wada Y. J Biol Chem. 1997;272:24530–24535. doi: 10.1074/jbc.272.39.24530. [DOI] [PubMed] [Google Scholar]
- 27.Bassham D C, Raikhel N V. Plant Physiol. 1998;117:407–415. doi: 10.1104/pp.117.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahmed S U, Bar-Peled M, Raikhel N V. Plant Physiol. 1997;114:325–336. doi: 10.1104/pp.114.1.325. , and addendum (1998) 115, 311–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paris N, Rogers S W, Jiang L, Kirsch T, Beevers L, Phillips T E, Rogers J C. Plant Physiol. 1997;115:29–39. doi: 10.1104/pp.115.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rapoport I, Miyazaki M, Boll W, Duckworth B, Cantley L C, Shoelson S, Kirchhausen T. EMBO J. 1997;16:2240–2250. doi: 10.1093/emboj/16.9.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rapoport I, Chen Y C, Cupers P, Shoelson S E, Kirchhausen T. EMBO J. 1998;17:2148–2155. doi: 10.1093/emboj/17.8.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maeshima M, Yoshida S. J Biol Chem. 1989;264:20068–20073. [PubMed] [Google Scholar]
- 33.Ueda T, Anai T, Tsukaya H, Hirara A, Uchimiya H. Mol Gen Genet. 1996;250:533–539. doi: 10.1007/BF02174441. [DOI] [PubMed] [Google Scholar]
- 34.Slot J W, Geuze H J, Gigengack S, Lienhard G E, James D E. J Cell Biol. 1991;113:123–135. doi: 10.1083/jcb.113.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffiths S, Simons K, Warren G, Tokuyasu K T. Methods Enzymol. 1983;96:466–485. doi: 10.1016/s0076-6879(83)96041-x. [DOI] [PubMed] [Google Scholar]
- 36.Ohno H, Stewart J, Fournier M C, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino J S. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- 37.Chrispeels M J, Raikhel N V. Cell. 1992;68:613–616. doi: 10.1016/0092-8674(92)90134-x. [DOI] [PubMed] [Google Scholar]
- 38.Paris N, Stanley C M, Jones R L, Rogers J C. Cell. 1996;85:563–572. doi: 10.1016/s0092-8674(00)81256-8. [DOI] [PubMed] [Google Scholar]
- 39.Shimada T, Kuroyanag M, Nishimura M, Hara-Nishimuram I. Plant Cell Physiol. 1997;38:1414–1420. doi: 10.1093/oxfordjournals.pcp.a029138. [DOI] [PubMed] [Google Scholar]