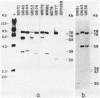
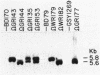
Abstract
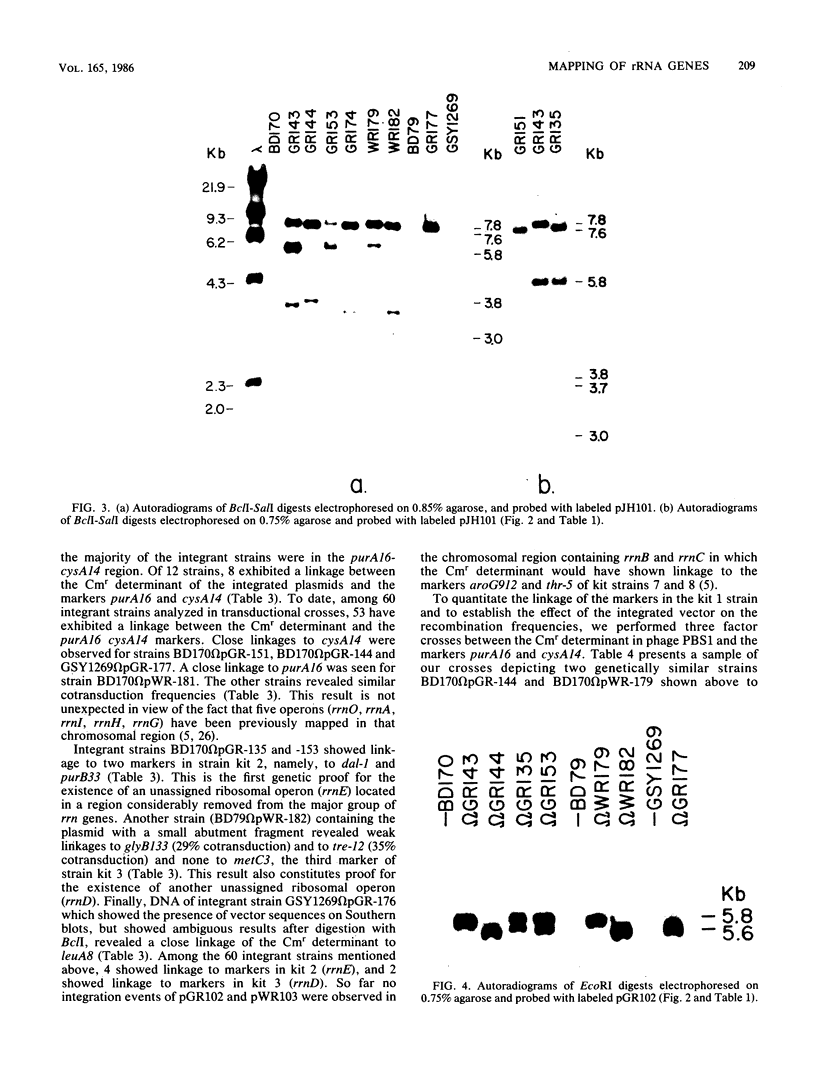
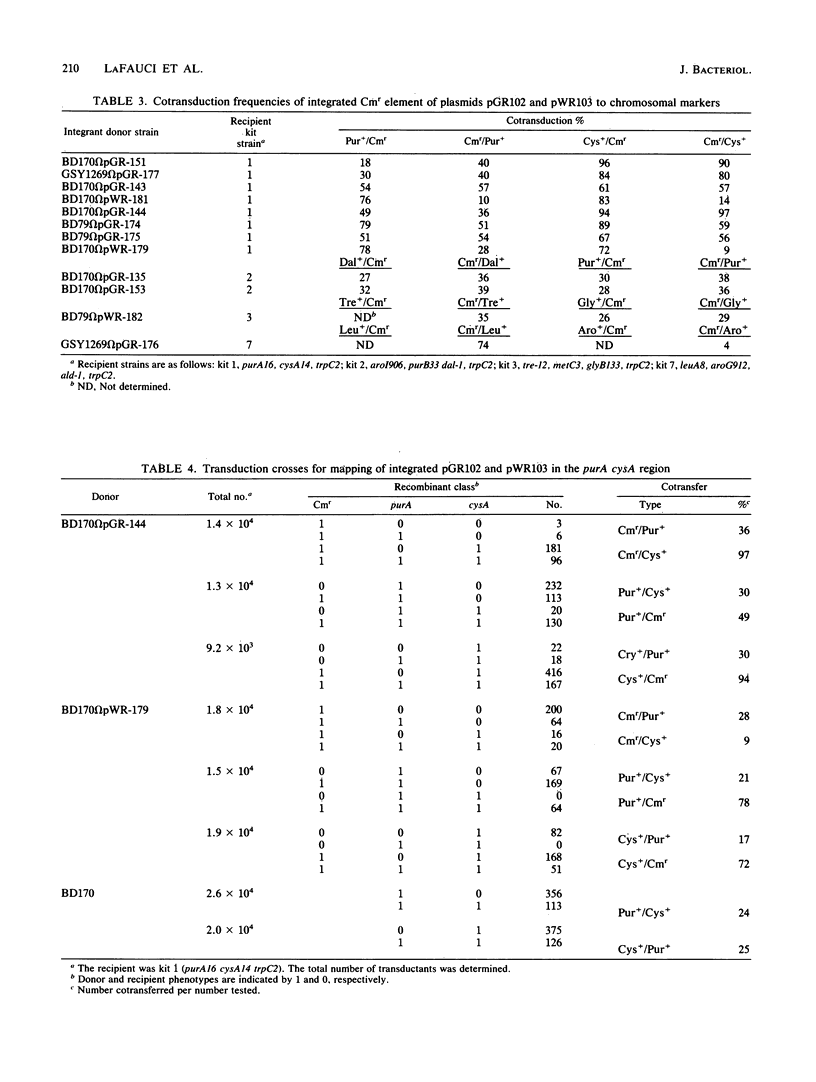
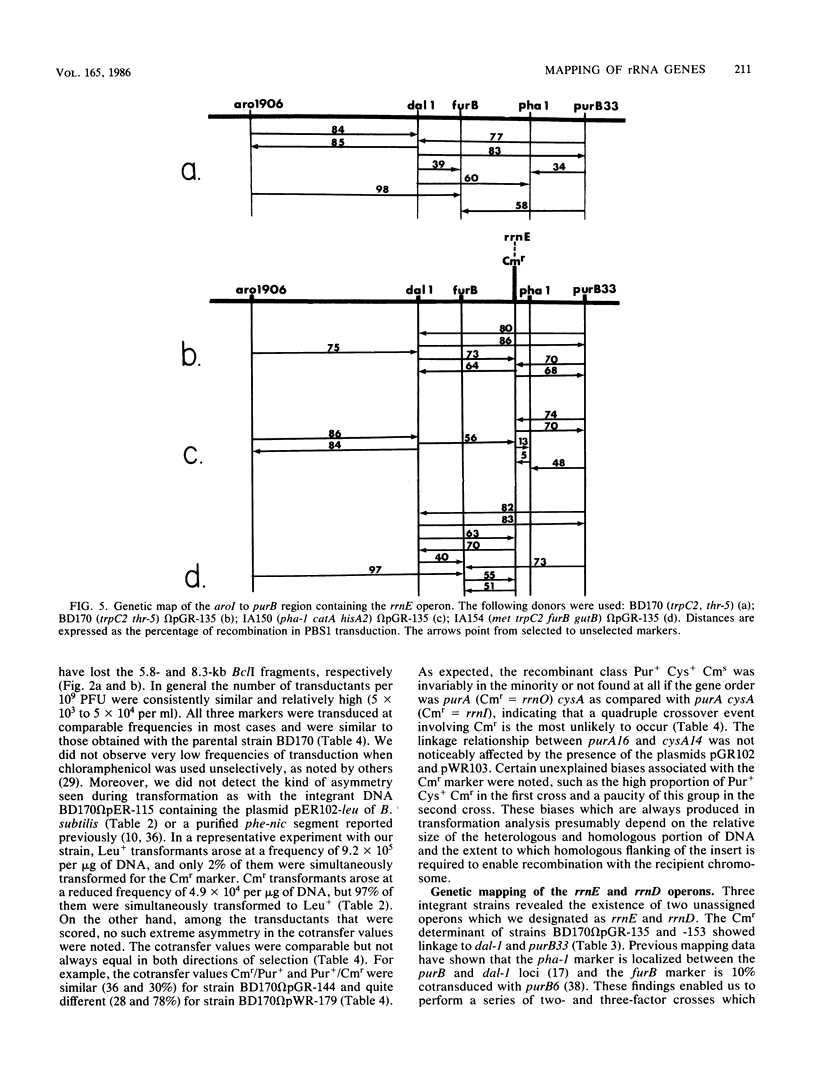
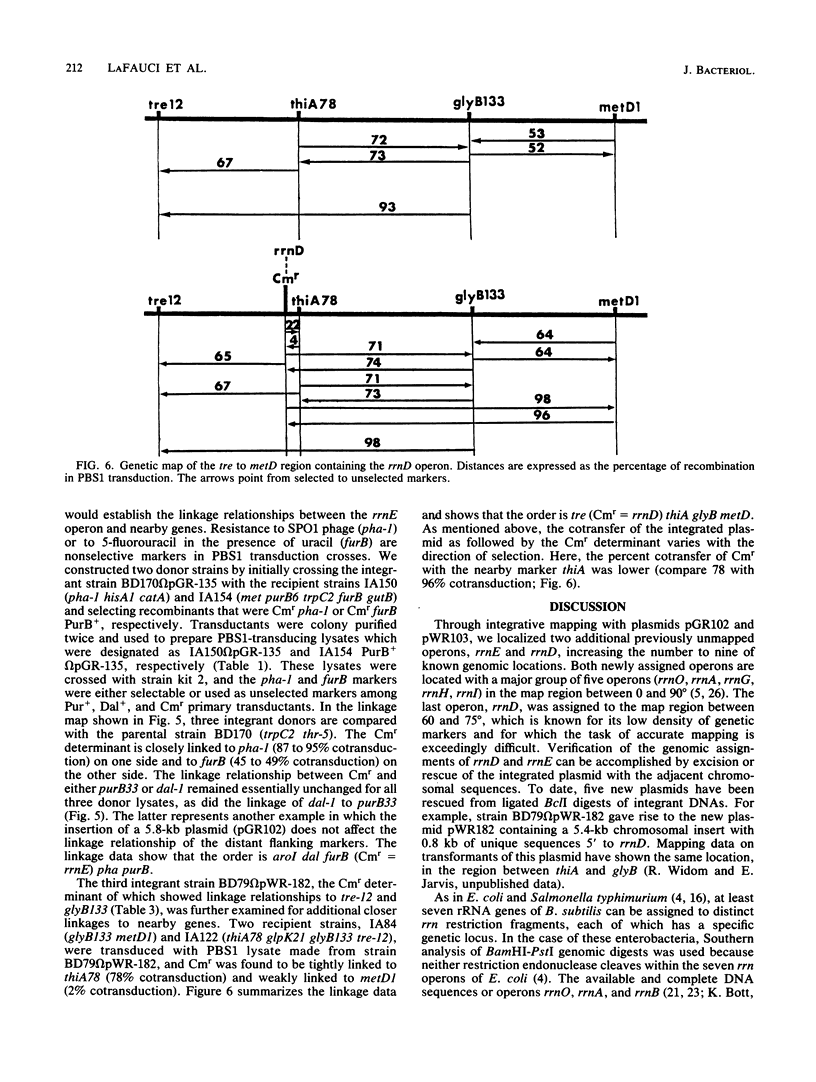
Integrable plasmids pGR102 and pWR103 containing ribosomal sequences from within the transcriptional units for 16S and 23S were used to transform Bacillus subtilis. To date, these plasmids integrated into 7 of 10 known rrn operons. Two such events occurred at unassigned operons, revealing a close linkage of the CAT gene of the plasmid to pha-1 situated between dal-1 and purB33 for rrnE and to thiA78 situated between glyB133 and re-12 for rrnD. All seven integration events that led to the loss of unique ribosomal BclI fragments can now be assigned to known rrn operons.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Galizzi A. Amplification of a chromosomal region in Bacillus subtilis. J Bacteriol. 1985 Jun;162(3):1203–1211. doi: 10.1128/jb.162.3.1203-1211.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros I., Kiss A., Venetianer P. Physical map of the seven ribosomal RNA genes of Escherichia coli. Nucleic Acids Res. 1979;6(5):1817–1830. doi: 10.1093/nar/6.5.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canosi U., Morelli G., Trautner T. A. The relationship between molecular structure and transformation efficiency of some S. aureus plasmids isolated from B. subtilis. Mol Gen Genet. 1978 Nov 9;166(3):259–267. doi: 10.1007/BF00267617. [DOI] [PubMed] [Google Scholar]
- Chilton M. D., McCarthy B. J. Genetic and base sequence homologies in bacillus. Genetics. 1969 Jul;62(3):697–710. [PMC free article] [PubMed] [Google Scholar]
- Dedonder R. A., Lepesant J. A., Lepesant-Kejzlarová J., Billault A., Steinmetz M., Kunst F. Construction of a kit of reference strains for rapid genetic mapping in Bacillus subtilis 168. Appl Environ Microbiol. 1977 Apr;33(4):989–993. doi: 10.1128/aem.33.4.989-993.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwood M., Nomura M. Deletion of a ribosomal ribonucleic acid operon in Escherichia coli. J Bacteriol. 1980 Aug;143(2):1077–1080. doi: 10.1128/jb.143.2.1077-1080.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F. A., Nguyen A., Lang D., Hoch J. A. Construction and properties of an integrable plasmid for Bacillus subtilis. J Bacteriol. 1983 Jun;154(3):1513–1515. doi: 10.1128/jb.154.3.1513-1515.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb P., LaFauci G., Rudner R. Alterations in the number of rRNA operons within the Bacillus subtilis genome. Gene. 1985;33(3):259–268. doi: 10.1016/0378-1119(85)90233-1. [DOI] [PubMed] [Google Scholar]
- Haldenwang W. G., Banner C. D., Ollington J. F., Losick R., Hoch J. A., O'Connor M. B., Sonenshein A. L. Mapping a cloned gene under sporulation control by inserttion of a drug resistance marker into the Bacillus subtilis chromosome. J Bacteriol. 1980 Apr;142(1):90–98. doi: 10.1128/jb.142.1.90-98.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J. A. Genetic analysis of pleiotropic negative sporulation mutants in Bacillus subtilis. J Bacteriol. 1971 Mar;105(3):896–901. doi: 10.1128/jb.105.3.896-901.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias A., Ceglowski P., Trautner T. A. Plasmid transformation in Bacillus subtilis. Effects of the insertion of Bacillus subtilis rRNA genes into plasmids. Mol Gen Genet. 1983;192(1-2):149–154. doi: 10.1007/BF00327660. [DOI] [PubMed] [Google Scholar]
- Lehner A. F., Harvey S., Hill C. W. Mapping and spacer identification of rRNA operons of Salmonella typhimurium. J Bacteriol. 1984 Nov;160(2):682–686. doi: 10.1128/jb.160.2.682-686.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepesant-Kejzlarová J., Lepesant J. A., Walle J., Billault A., Dedonder R. Revision of the linkage map of Bacillus subtilis 168: indications for circularity of the chromosome. J Bacteriol. 1975 Mar;121(3):823–834. doi: 10.1128/jb.121.3.823-834.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughney K., Lund E., Dahlberg J. E. Deletion of an rRNA gene set in Bacillus subtilis. J Bacteriol. 1983 Apr;154(1):529–532. doi: 10.1128/jb.154.1.529-532.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughney K., Lund E., Dahlberg J. E. tRNA genes are found between 16S and 23S rRNA genes in Bacillus subtilis. Nucleic Acids Res. 1982 Mar 11;10(5):1607–1624. doi: 10.1093/nar/10.5.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya S., Ogasawara N., Yoshikawa H. Structure and function of the region of the replication origin of the Bacillus subtilis chromosome. III. Nucleotide sequence of some 10,000 base pairs in the origin region. Nucleic Acids Res. 1985 Apr 11;13(7):2251–2265. doi: 10.1093/nar/13.7.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahari K., Sakaguchi K. Cloning of Bacillus subtilis leucina A, B and C genes with Escherichia coli plasmids and expression of the leuC gene in E. coli. Mol Gen Genet. 1978 Jan 17;158(3):263–270. doi: 10.1007/BF00267197. [DOI] [PubMed] [Google Scholar]
- Ogasawara N., Moriya S., Yoshikawa H. Structure and organization of rRNA operons in the region of the replication origin of the Bacillus subtilis chromosome. Nucleic Acids Res. 1983 Sep 24;11(18):6301–6318. doi: 10.1093/nar/11.18.6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara N., Seiki M., Yoshikawa H. Replication origin region of Bacillus subtilis chromosome contains two rRNA operons. J Bacteriol. 1983 Apr;154(1):50–57. doi: 10.1128/jb.154.1.50-57.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostapchuk P., Anilionis A., Riley M. Conserved genes in enteric bacteria are not identical. Mol Gen Genet. 1980;180(2):475–477. doi: 10.1007/BF00425864. [DOI] [PubMed] [Google Scholar]
- Piggot P. J., Hoch J. A. Revised genetic linkage map of Bacillus subtilis. Microbiol Rev. 1985 Jun;49(2):158–179. doi: 10.1128/mr.49.2.158-179.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rudner R., Lin H. J., Hoffmann E. M., Chargaff E. Studies on the loss and the restoration of the transforming activity of the deoxyribonucleic acid of Bacillus subtilis. Biochim Biophys Acta. 1967 Nov 21;149(1):199–219. doi: 10.1016/0005-2787(67)90702-2. [DOI] [PubMed] [Google Scholar]
- Sargent M. G., Bennett M. F. Amplification of a major membrane-bound DNA sequence of Bacillus subtilis. J Bacteriol. 1985 Feb;161(2):589–595. doi: 10.1128/jb.161.2.589-595.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiki M., Ogasawara N., Yoshikawa H. Structure and function of the region of the replication origin of the Bacillus subtilis chromosome. I. Isolation and characterization of plasmids containing the origin region. Mol Gen Genet. 1981;183(2):220–226. doi: 10.1007/BF00270621. [DOI] [PubMed] [Google Scholar]
- Smith I., Dubnau D., Morrell P., Marmur J. Chromosomal location of DNA base sequences complementary to transfer RNA and to 5 s, 16 s and 23 s ribosomal RNA in Bacillus subtilis. J Mol Biol. 1968 Apr 14;33(1):123–140. doi: 10.1016/0022-2836(68)90285-4. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stewart G. C., Wilson F. E., Bott K. F. Detailed physical mapping of the ribosomal RNA genes of Bacillus subtilis. Gene. 1982 Sep;19(2):153–162. doi: 10.1016/0378-1119(82)90001-4. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Weisblum B. Construction of a colicin E1-R factor composite plasmid in vitro: means for amplification of deoxyribonucleic acid. J Bacteriol. 1975 Jan;121(1):354–362. doi: 10.1128/jb.121.1.354-362.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. Gene amplification in Bacillus subtilis. J Gen Microbiol. 1984 Jul;130(7):1613–1621. doi: 10.1099/00221287-130-7-1613. [DOI] [PubMed] [Google Scholar]
- Young M. The mechanism of insertion of a segment of heterologous DNA into the chromosome of Bacillus subtilis. J Gen Microbiol. 1983 May;129(5):1497–1512. doi: 10.1099/00221287-129-5-1497. [DOI] [PubMed] [Google Scholar]