Abstract
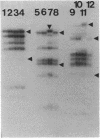
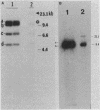
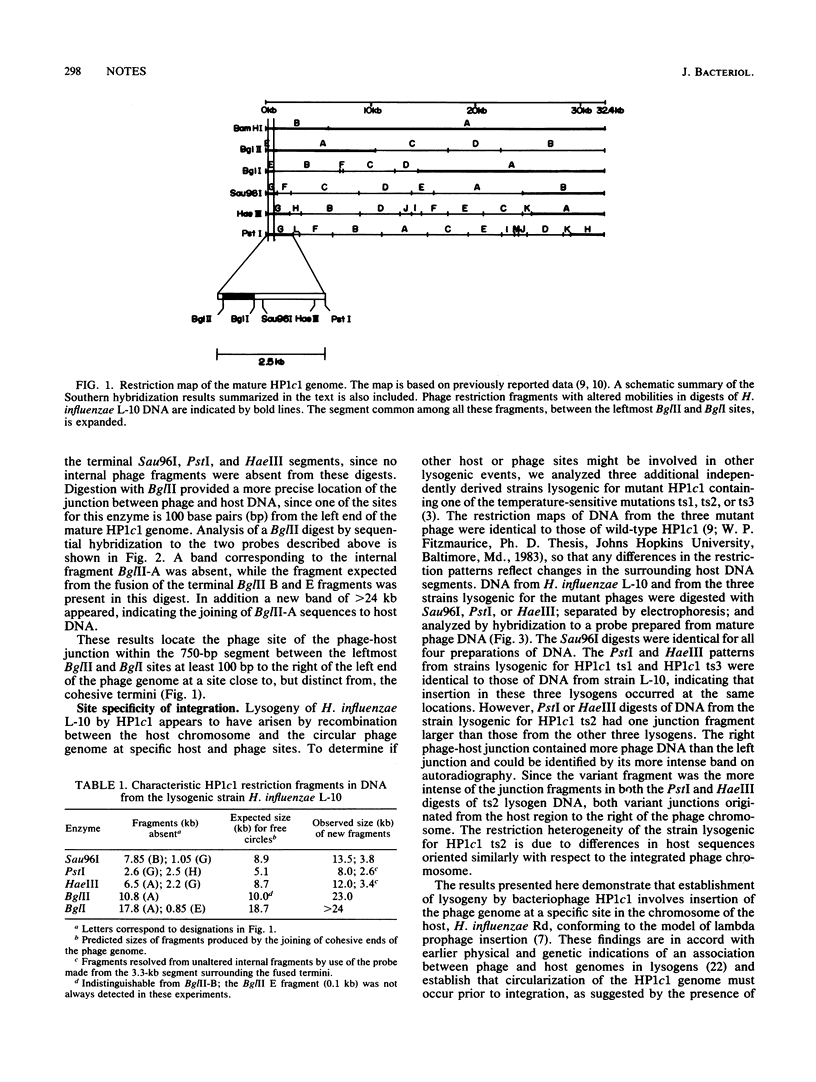
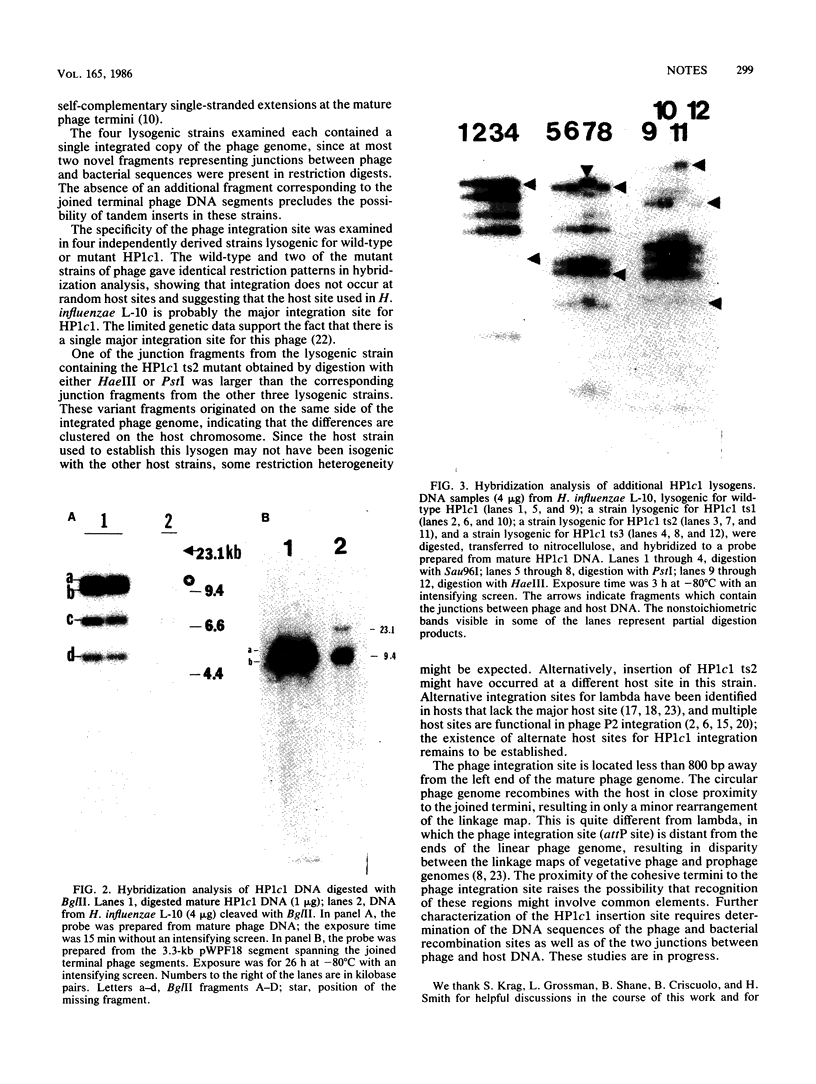
Restriction fragments hybridizing to phage HP1c1 DNA were identified in digests of DNA from lysogenic strains of Haemophilus influenzae. The results showed that the cohesive ends of the mature phage DNA were joined in lysogens and that the phage genome was covalently linked to the host DNA, indicating that lysogeny involves recombination between specific sites on the phage and host chromosomes. The site on the phage chromosome at which this recombination occurred was between 110 and 750 base pairs of the left end on the mature phage genome.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BERTANI G., SIX E. Inheritance of prophage P2 in bacterial crosses. Virology. 1958 Oct;6(2):357–381. doi: 10.1016/0042-6822(58)90089-8. [DOI] [PubMed] [Google Scholar]
- Benjamin R. C., Fitzmaurice W. P., Huang P. C., Scocca J. J. Nucleotide sequence of cloned DNA segments of the Haemophilus influenzae bacteriophage HP1c1. Gene. 1984 Nov;31(1-3):173–185. doi: 10.1016/0378-1119(84)90208-7. [DOI] [PubMed] [Google Scholar]
- Boling M. E., Setlow J. K. Dependence of Vegetative Recombination Among Haemophilus influenzae Bacteriophage on the Host Cell. J Virol. 1969 Sep;4(3):240–243. doi: 10.1128/jvi.4.3.240-243.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari A. I., Metlay M. Genetic mapping of prophage Mu. Virology. 1973 Jul;54(1):109–116. doi: 10.1016/0042-6822(73)90120-7. [DOI] [PubMed] [Google Scholar]
- Fitzmaurice W. P., Scocca J. J. Restriction map and location of mutations on the genome of bacteriophage Hp1c1 of Haemophilus influenzae Rd. Gene. 1983 Sep;24(1):29–35. doi: 10.1016/0378-1119(83)90128-2. [DOI] [PubMed] [Google Scholar]
- Fitzmaurice W. P., Waldman A. S., Benjamin R. C., Huang P. C., Scocca J. J. Nucleotide sequence and properties of the cohesive DNA termini from bacteriophage HP1c1 of Haemophilus influenzae Rd. Gene. 1984 Nov;31(1-3):197–203. doi: 10.1016/0378-1119(84)90210-5. [DOI] [PubMed] [Google Scholar]
- HARM W., RUPERT C. S. INFECTION OF TRANSFORMABLE CELLS OF HAEMOPHILUS INFLUENZAE BY BACTERIOPHAGE AND BACTERIOPHAGE DNA. Z Vererbungsl. 1963 Dec 30;94:336–348. doi: 10.1007/BF00897593. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Hsu M. T., Davidson N. Structure of inserted bacteriophage Mu-1 DNA and physical mapping of bacterial genes by Mu-1 DNA insertion. Proc Natl Acad Sci U S A. 1972 Oct;69(10):2823–2827. doi: 10.1073/pnas.69.10.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Tomizawa J. Prophage P1, and extrachromosomal replication unit. Cold Spring Harb Symp Quant Biol. 1968;33:791–798. doi: 10.1101/sqb.1968.033.01.091. [DOI] [PubMed] [Google Scholar]
- KELLY B. Localization of P2 prophage in two strains of Escherichia coli. Virology. 1963 Jan;19:32–39. doi: 10.1016/0042-6822(63)90021-7. [DOI] [PubMed] [Google Scholar]
- Lindahl G. Multiple recombination mechanisms in bacteriophage P2. Virology. 1969 Dec;39(4):861–866. doi: 10.1016/0042-6822(69)90022-1. [DOI] [PubMed] [Google Scholar]
- Pinkham J. L., Platt T., Enquist L. W., Weisberg R. A. The secondary attachment site for bacteriophage lambda in the proA/B gene of Escherichia coli. J Mol Biol. 1980 Dec 25;144(4):587–592. doi: 10.1016/0022-2836(80)90339-3. [DOI] [PubMed] [Google Scholar]
- Ross W., Landy A. Patterns of lambda Int recognition in the regions of strand exchange. Cell. 1983 May;33(1):261–272. doi: 10.1016/0092-8674(83)90355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scocca J. J., Poland R. L., Zoon K. C. Specificity in deoxyribonucleic acid uptake by transformable Haemophilus influenzae. J Bacteriol. 1974 May;118(2):369–373. doi: 10.1128/jb.118.2.369-373.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stuy J. H. Transfer of genetic information within a colony of Haemophilus influenzae. J Bacteriol. 1985 Apr;162(1):1–4. doi: 10.1128/jb.162.1.1-4.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeldis J. B., Bukhari A. I., Zipser D. Orientation of prophage Mu. Virology. 1973 Sep;55(1):289–294. doi: 10.1016/s0042-6822(73)81033-5. [DOI] [PubMed] [Google Scholar]