Abstract
The human β-globin locus is activated transcriptionally by a complex series of events that culminate in appropriate temporal and tissue-specific control over five separate genes during embryonic and early postnatal development. One cis-regulatory element in the locus, originally identified as an enhancer 3′ to the Aγ-globin gene, more recently has been suggested to harbor alternative or additional properties, including stage-specific silencer, insulator, nuclear matrix, or chromosome scaffold attachment activities. We have re-evaluated the activity during erythropoiesis that is conferred by this element by deleting it from a yeast artificial chromosome (YAC) containing the entire human β-globin locus and then assaying for the expression of each gene at each developmental stage after incorporation of the mutant YAC into the mouse germline. The data show that loss of the Aγ-globin 3′ element confers no phenotype in six independent lines of intact YAC mutant transgenic mice, thus demonstrating (minimally) that any activities attributable to this element are fully compensated by other DNA sequences within the β-globin locus.
At least 17 discrete transcriptional control elements lie within the human β-globin gene locus. These include the five DNaseI hypersensitive sites lying from 6 to 24 kbp 5′ to the ɛ-globin gene [the locus control region (LCR) (1–3)], the promoters of each of five separate genes, and distinct modulatory elements that lie either within, or in relatively close proximity to, one or another of the genes.
Among these gene-proximal regulatory elements, one of the earliest to be identified was an enhancer element discovered over a decade ago in transient transfection experiments (4). This sequence lies 410 bp 3′ to the Aγ-globin gene and is confined within boundaries originally described by a 757-bp EcoRI/HindIII fragment. With passing time, however, the bona fide regulatory activity conferred by this element became more controversial because subsequent studies in transgenic mice implicated this same element in the stage-specific and autonomous silencing of the Aγ-globin gene in adult stage erythroid cells (5, 6). Subsequently, in vitro studies implicated the same element as a chromatin structural motif that could participate in tethering the locus to chromosome scaffolds or to the nuclear matrix (7). More recently, it was shown that this sequence, in collaboration with the LCR, is able to shelter γ-globin transgenes from variegation effects caused by the neighboring chromatin into which they randomly integrate, suggesting that this combination of cis-elements can provide insulation from the repressive effects of surrounding heterochromatin (8). However, even the most current analyses demonstrate that the activity due to this sequence is still controversial and unresolved (9).
These multiple observations and seemingly disparate conclusions then provoke several questions: Is this Aγ-globin 3′ element (referred to hereafter as the Aγ3′E) an enhancer, a silencer, or a sequence required for appropriate chromatin configuration or activity within the erythroid cell nucleus, or could it perhaps serve all of these functions at different stages during erythropoiesis? Although several different methods have been used to address the activity of this element in previous reports (in transient or stable transfection studies as well as in transgenic animal models), all of them used specifically selected segments of the human globin gene locus for analysis.
We previously determined that the activities of selected fragments artificially brought together by ligation into a single plasmid are often inconsistent with the regulatory properties that are elicited after elimination of specific cis-elements in the context of the intact β-globin locus (10, 11). We therefore examined the activity of this regulatory sequence directly by deleting the Aγ3′E from the intact locus borne on a yeast artificial chromosome (YAC) and then examined the regulatory response in animals bearing the mutant transgenes for any alterations in normal human β-globin gene expression. The results of the analysis were clear: There is no alteration in the timing or level of expression of any of the genes within the locus at any stage of erythropoiesis, indicating either that the activity of the Aγ3′E is fully compensated by other sequences within the locus or that it has no regulatory activity.
MATERIALS AND METHODS
Targeting Constructs.
To generate the target constructs for homologous recombination in yeast, we amplified two PCR fragments corresponding to the 5′ flanking and 3′ flanking regions of the Aγ3′E and ligated these two fragments into the HindIII/BamHI site of pRS306. An 800-bp fragment corresponding to the 5′ flanking region of the Aγ3′E was amplified by PCR from genomic DNA, introducing the restriction enzyme sites HindIII and XhoI at the 5′ and 3′ ends, respectively (primer sequences were: γ5′-FU ACGTAAGCTTAGAGTAGCAAGATTTAAATTA, and γ5′-FD GATACTCGAGGGAACACTTTCCCTTCATTA, where the underlined nucleotides correspond to position 40,561 and 41,364 of the human β-globin locus sequence. At Web site: http://globin.cse.psu.edu). The 1,600-bp 3′ flanking sequence also was generated by PCR, introducing XhoI and BamHI sites on the 5′ and 3′ ends of that fragment, respectively (using primers γ3′-FU CATGCTCGAGGGGTTTTGAGTGAACTACAG and γ3′-FD ATCGGGATCCCGTACCAGTGAACAGTTGCC, where the two underlined nucleotides indicate positions 42,121 and 43,771 of the globin locus).
Generation of the Aγ-Globin 3′ Element Deletion YAC Mutant.
The YAC mutant was generated by a two-step procedure involving site-specific integration of the pRS306 targeting construct, harboring the homologous sequences flanking the Aγ 3′ enhancer, into the human β-globin YAC A201F4.2, followed by excision of the wild-type copy (together with the URA3 marker gene). Both events were mediated by yeast homologous recombination. After transformation of the yeast clone A201F4.2 with the URA3 targeting vector (linearized with EcoRI), yeast colonies were plated onto selective medium (Lys-, Trp-, Ura-). DNA was isolated from individual colonies and subjected to restriction analysis and Southern blotting to identify those clones that displayed the correct integration pattern (not shown). These clones were transferred to Lys-, Trp-, and Ura+ media to allow reversion, which excises either the wild-type or the mutant integrant plasmid from the locus. Removal of the Aγ3′E, and determination of the structural integrity of the YACs after homologous recombination, was verified by normal or pulse-field gel electrophoresis (PFGE) and Southern blot hybridizations.
Transgenic Mice: Integrated Transgene Characterization.
YAC DNA was purified after PFGE (10) and injected into fertilized mouse eggs (CD1; Charles River Breeding Laboratory). Oocytes surviving the injections were transferred to foster mothers as described (12). Tail DNAs from offspring were analyzed for the presence of the left and right YAC vector arms by PCR (11). Transgene copy number then was assessed by Southern blot hybridization as described (11), and the copy numbers for lines 1 through 6, respectively, were 2, 1, 1, 4, 1, and 2. To assess the integrity of the integrated YAC transgenes, cells recovered from 0.15 g of transgenic thymus were diluted in 1 ml of cell suspension buffer (10 mM Tris⋅HCl, pH 7.5/20 mM NaCl/50 mM EDTA, pH 8.0). The cell solution then was diluted to 0.4% in low melting preparative agarose and poured into clamped homogeneous electric field plug molds. Solidified plugs then were treated at 50°C in Proteinase K buffer (100 mM EDTA, pH 8.0/0.2% sodium deoxycholate/1% sarkosyl/1 mg/ml proteinase K) for 48 h, followed by several 1-h washes with 10 mM Tris⋅HCl (pH 7.0), 1 mM EDTA buffer. The plugs then were digested with SfiI at 50°C for 12 h, melted at 65°C for 20 min, and loaded into pulsed-field gel wells. The gels were electrophoresed and then blotted to nylon filters (11). Four radiolabeled probes were used [corresponding to HS3, the ɛ- and β-globin genes, and downstream sequence RK29 (13)], which should all, if the YAC had integrated intact into the genome, hybridize to identical SfiI fragments (each >97 kbp) in each transgenic line.
Semiquantitative Multiplex Reverse Transcriptase-PCR Assay.
The assay was developed and described in detail elsewhere (10), with a modification replacing the previously used γ-globin primer pair with a new set of primers that span an intron. The sequence of the new γ primers was: hγs: 5′-GATGCCATAAAGCACCTGGATG-3′ and hγas: 5′-TTGCAGAATAAAGCCTATCCTTGA-3′, where the underlined nucleotides correspond to positions 34,893 and 36,007 in the globin sequence database (above). A titration of the cycle number was performed on each cDNA sample to verify that the amplification of all templates was within the linear range. An aliquot of each PCR was electrophoresed on 8% polyacrylamide gels, dried, and subjected to autoradiography. Individual band intensities were quantified on a PhosphorImager (Molecular Dynamics) (14); expression per gene copy (above) was calculated as described (10). Two samples from each developmental stage in erythropoiesis (9.5 dpc yolk sac, 14.5 dpc fetal liver or anemic adult spleen) were analyzed from multiple siblings, RNA preparations, and different litters for each transgenic line.
RESULTS
Deletion of the Aγ3′ Element from a Human β-Globin YAC.
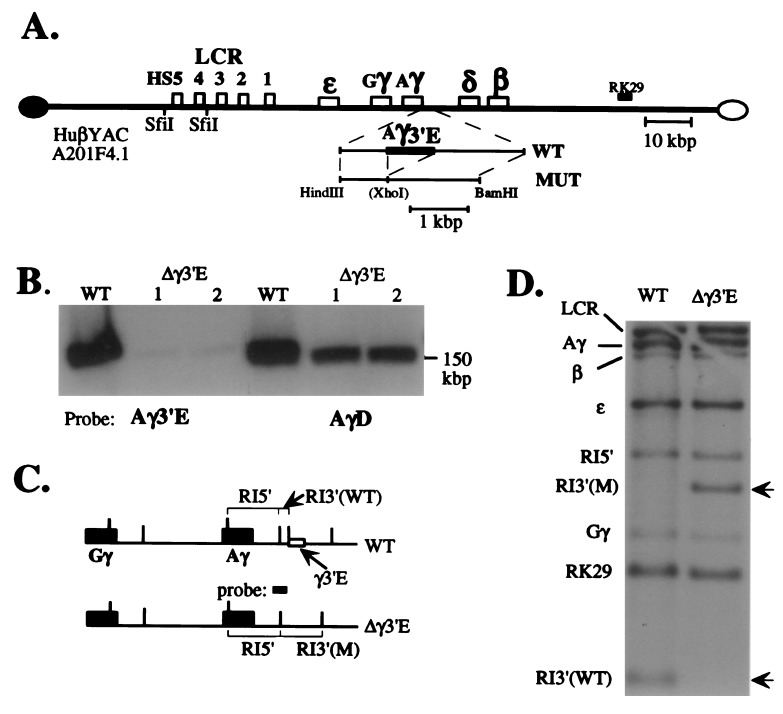
Two human β-globin locus YACs have been shown to regulate all of the genes in proper tissue- and stage-specific manner when they are incorporated into the mouse germline (10, 13, 15, 16). The Aγ3′E was deleted from the 150-kbp YAC by using homologous recombination in yeast (17), as described in detail (10). The mutagenesis strategy and the structure of both the wild-type and mutated YACs are described in Fig. 1. Fig. 1A is a diagrammatic representation of the predicted structure of human β-globin YAC A201F4.2 (10, 13) before and after deletion of the 757-bp Aγ3′E (4). To verify that the mutant depicted in Fig. 1A was, in fact, the structure recovered in the YAC used for microinjection, the mutant YAC was characterized structurally by PFGE and Southern blotting experiments. These data showed that the mutant YAC is unaltered in overall size but no longer contains the Aγ3′E (Fig. 1B). To confirm that the detailed structure of the rest of the locus was unaltered during these manipulations, yeast genomic DNA, prepared from both the wild-type and mutant YAC strains, was digested with EcoRI (recognition sites indicated by vertical lines in Fig. 1C), transferred to nylon filters, and hybridized to radiolabeled probes spanning the locus [corresponding to the LCR, the ɛ-, γ- and β-globin genes, as well as the 3′ flanking marker, RK29 (13)]. All of the restriction fragments analyzed were of the predicted size (Figs. 1 C and D), indicating that the overall structure of the YAC was not altered by the manipulations. To demonstrate that the mutation specifically deleted the Aγ3′E, the same filter was re-hybridized with a probe overlapping the 5′ flanking region of the deletion (Fig. 1C). The deletion resulted in the specific loss of a 0.4-kbp EcoRI fragment from the wild-type YAC and creation of a new 2.1-kbp EcoRI mutant fragment, as predicted (Fig. 1D, arrowheads).
Figure 1.
Generation and characterization of the 3′ γ-globin element deletion mutant YAC. (A) Diagrammatic representation of wild-type and mutant YACs. An expanded diagram depicting the targeting construct generated here (abbreviated MUT) is shown below the diagram of the wild-type YAC (the position of the Aγ-3′ element deletion is indicated by a fusion within the MUT subclone). The position of the probes used in Fig. 2 (below) are indicated. (B) Wild-type and two separate subclones of the mutant YAC DNAs (1 and 2) were separated from yeast chromosomes by PFGE, transferred to nylon filters, and hybridized to either the Aγ 3′ element (Aγ3′E) or to a sequence lying 3′ to the Aγ3′E (called “AγD”; nucleotides 44,350–44,750 of the locus; www.globin.cse.psu.edu) to verify that the Aγ3′E was deleted specifically and that the gross size of the mutant YAC was not altered during the manipulations (PFGE does not resolve the difference in size between the two 150-kbp YACs ± the 757-bp Aγ3′E). Faint hybridization is detected in the Aγ3′E deletion mutant lanes with the Aγ3′E probe due to homology with the corresponding Gγ sequence (27), which shows no biological activity (4). (C) A diagram depicting both the wild-type and predicted mutant YAC structures in the vicinity of the γ-globin genes. Recognition sites for EcoRI are depicted as vertical lines. The “probe” (spanning the 3′ gene-proximal EcoRI site) referred to here and in D was generated from the genomic locus from nucleotides 40,861–41,867. (D) A Southern blot showing that the wild-type and Aγ3′E mutant YACs bear identical EcoRI fragments hybridizing to the LCR (10.4 kbp), RK29 (1.0 kbp), ɛ- (3.7 kbp), γ- (6.9 kbp), and β-globin (5.5 kbp) genes (see Fig. 2A). Two different EcoRI fragments hybridize to a probe spanning part of the deletion (C). Note that the Aγ3′E deletion results in the loss of a 0.4-kbp EcoRI fragment, simultaneously creating a 2.1-kbp EcoRI fragment. This probe (C) also crosshybridizes with a Gγ-globin 3′ fragment, 1.6 kbp in size.
Structural Integrity of the Mutant Human β-Globin YAC in Transgenic Mice.
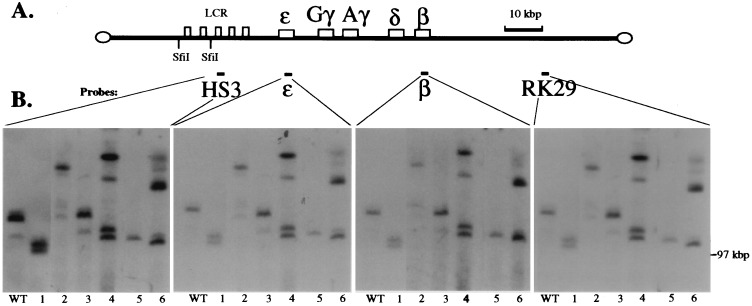
We next generated transgenic lines carrying the YAC bearing the deletion of the Aγ3′E. We generated 10 transgenic founder animals bearing the mutant YAC; subsequent analyses demonstrated that, of these founders, only six contained transgenes that both transmitted to progeny and bore mutant YACs that were physically intact (below). The structural integrity and copy number of these YAC transgenes was determined by PFGE/Southern hybridization (11). High molecular mass DNA, isolated from the thymuses of each of these six transgenic lines as well as from a single copy wild-type line (10), was digested with SfiI, transferred to nylon filters, and hybridized to probes corresponding to HS3, ɛ-globin, β-globin, or RK29 (Figs. 2 A and B). The fact that all of the probes spanning the locus hybridize to identical SfiI fragments in each of the transgenic lines and that each of these hybridizing bands is larger than 100 kbp (the distance from the SfiI site between HS3 and HS4 to the 3′ end of the YAC) indicates that all of these YAC transgenes are substantially intact (Fig. 2B). According to YAC end analysis (ref. 11 and data not shown) and additional Southern mapping assays, we determined that the six transgenic lines examined here contained from one to four intact YAC transgene copies (Fig. 2).
Figure 2.
Mapping the integrated Aγ3′E YAC transgene structures. (A) Diagram of the human β-globin YAC showing the SfiI sites and probes used. (B) Analysis of the integrity of the YAC transgenic lines. High molecular mass DNA isolated from thymus cells of transgenic lines was digested with SfiI and subjected to PFGE (see Materials and Methods). The gel was blotted to nylon filters and hybridized to probes corresponding to HS3, ɛ- and β-globin genes, and 3′ flanking marker RK29, respectively, in the four panels shown. The position of the bacteriophage λ dimer marker run in parallel (97 kbp) is shown on the right. All four probes hybridize to SfiI fragments larger than 100 kbp, and each probe hybridizes to the identical bands in each transgenic line.
Lack of the Aγ3′E Does Not Alter Any Aspect of Human β-Globin Locus Gene Expression.
The expression profile of the individual human globin genes during erythroid development was examined in each transgenic line by using a semi-quantitative reverse transcriptase-PCR assay that was developed and described previously (10), including in each PCR four sets of primers that uniquely amplify the human ɛ-, both γ-, and the adult β-globin genes, as well as the murine α-globin gene, which serves as an internal control. (However, we used a new set of primers for the γ-globin gene that now also spans an intron; see Materials and Methods). Multiple, independent RNA samples were recovered from each of the transgenic lines at 9.5 dpc (yolk sac) and 14.5 dpc (fetal liver) and from anemic adult spleens as representative sites and times of maximal human ɛ-, γ-, and β-globin synthesis during mouse erythroid development, respectively (15). The RNA samples then were reverse transcribed by using random primers and amplified to mid-exponential phase by using the globin-specific primers, and finally the products were resolved by electrophoresis on 8% native polyacrylamide gels (10) and quantified on a PhosphorImager.
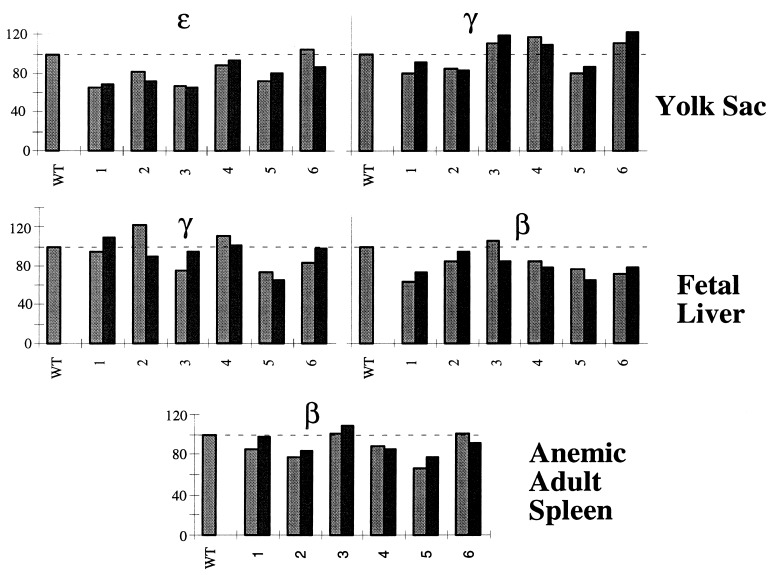
The results of this analysis revealed that the expression of all of the human β-locus genes in the Aγ-element mutant animals is remarkably similar to that observed in transgenic lines carrying the wild-type β-globin locus. As is apparent from the data, there is no compromise with respect to the stage of erythropoiesis at which each gene is expressed; thus, normal developmental control over stage-specificity of all genes, including the γ-globins, remains intact in the Aγ3′E mutant animals. Quantitative analysis of expression for two independent samples representing each time point in each of the transgenic lines (Fig. 3) revealed that all of the globin genes are expressed at normal abundance at every stage of development. This is in stark contrast, for example, to a LCR “core element” deletion mutant transgenic analysis, in which an extreme reduction in the expression of all of the genes was observed in single copy YAC transgenes missing either HS3 or HS4 (10). Similarly, deletion of either the minimal ɛ-globin “silencer” or the adult β-globin gene 3′ enhancer from the same YAC resulted in profound diminution in transcription of those two genes (11). We conclude from the data presented here that the deletion of the Aγ3′E does not affect high level, stage-specific, position-independent expression of any of the human globin genes in transgenic mice.
Figure 3.
Developmental expression of the Aγ3′E mutant YAC in transgenic mice. A semiquantitative reverse transcriptase-PCR assay was performed as described (10) to measure the expression of the human β-globin genes at each stage of erythroid development (see Materials and Methods); each bar represents an independent RNA sample. The radioactive PCR products for the mouse α-globin (internal control) and human ɛ-, γ-, and β-globin transcripts were separated on polyacrylamide gels and then quantified on a PhosphorImager. Ratios of hɛ/mα, hγ/mα, and hβ/mα expression per integrated transgene copy (see Materials and Methods) obtained from each mutant transgenic line at the three different developmental stages were compared with those from a single copy wild-type YAC transgenic mouse (taken as 100%).
DISCUSSION
In this study, we examined the effect of deleting the Aγ-globin 3′ element from a human β-globin YAC on the expression of all the human β-locus genes during erythroid development in transgenic mice. This region 3′ to the Aγ-globin gene has been the subject of intensive investigation, and a variety of different activities has been implicated in association with this cis element.
The 757-bp region that was deleted from the YAC in the present study is located 410 bp 3′ to the Aγ-globin gene polyadenylation site. Groudine et al. (18) originally mapped two DNaseI hypersensitive sites to this region. These sites were detected in human erythroid cell lines but were not found in nonerythroid cells, thus suggesting that these tissue-specific, hypersensitive sites might somehow be involved in the regulation of the β-globin locus. Subsequent studies identified this same sequence as an enhancer element, which was able to increase transcription levels of a reporter gene after transient transfection into both erythroid and nonerythroid cell lines (4). However, later studies examining various fragments of the locus reported that this same sequence possessed silencer activity (6) or indeed had no effect at all (19). Although yet a fifth analysis implicated the region in matrix or chromosome scaffold attachment (7), most recently a study examining the activity of various μLCR/Aγ-globin constructs in the presence or absence of the Aγ3′E concluded that it functions as a chromatin insulator, which, in collaboration with the LCR, confers position-independent expression to the γ-globin genes in transgenic mice (8, 20). In this respect, it is noteworthy that there are two high affinity, AT-rich binding sites for the scaffold attachment protein SATB1 within the Aγ3′E, suggesting that the element might provide an anchoring point for the locus to the nuclear matrix (21–23). The present results, however, show that deleting the Aγ3′E does not affect the position-independent expression of the globin genes. We conclude that the Aγ3′E is not important for conferring position-independent expression to the globin genes when analyzed within the context of the entire globin locus, thus suggesting that the “sheltering” or insulating activity attributed to the element may represent a redundant function that normally is compensated by other elements present in the locus.
These results clearly demonstrate that deletion of the Aγ 3′ element has no effect on the high level, stage-specific or position-independent expression of any of the human globin genes when analyzed in the context of the whole β-globin gene locus in transgenic mice, in substantial agreement with a preliminary report showing that deletion of the entire 12.5-kbp region lying between the human Aγ and δ globin genes results in no switching phenotype (24). We believe that these results once more underscore the importance of analyzing regulatory DNA elements in the context of the whole β-globin locus. There is a very large body of literature that has examined the role of individual regulatory DNA elements in the human globin gene locus where specific sequences were linked together in cis to various reporter genes and analyzed either in transfection studies or in transgenic mice. In several instances, these data have led to contradictory results concerning the functional role and significance of specific regulatory sequences. Accumulating evidence on other loci as well as the β-globin gene locus suggest that such elements, especially when situated within a complex regulatory environment, do not usually act alone but more often act in concert with other resident elements that act to either enhance, counteract, or compensate for the loss of any specific function.
In the present analysis, we did not observe either a decrease or increase in expression of any of the human β-globin genes after deletion of the Aγ3′E, as would have been predicted by deleting an enhancer or silencer (or chromatin modulatory) element. Several explanations could account for this observation. First, as alluded to above, it is possible that other, currently undefined, DNA elements can compensate for the loss of the Aγ3′E when its loss is assayed within the context of the whole globin gene locus. Alternatively, it should be kept in mind that the human globin genes, when introduced as transgenes into mice, are expressed in a similar stage-specific manner to the endogenous mouse β-globin genes (10, 15, 16, 25). Due reflection might be particularly important in this example (in which the mouse physiological model is being used for analysis of the human γ-globin genes) because mice do not exhibit a unique fetal stage of expression (26). During hematopoietic development in transgenic mice, the human γ-globin genes are expressed at very high levels in the embryonic yolk sac and are much less abundantly transcribed after hematopoiesis switches to the fetal liver. It is thus conceivable that the Aγ3′E contributes to changes in the stage-specific expression of the γ-globin genes only during human development, and thus, the evolutionary difference in mice and humans in erythropoiesis (i.e., displaying one genuine “switch” in synthesis, rather than two, respectively) might therefore account for any unique, uncompensated activity.
Acknowledgments
We thank Ms. Jie Fan for outstanding technical support and gratefully acknowledge fellowship support from the Cooley’s Anemia Foundation (Q.L.) and the Chicago chapter of the American Heart Association (J.B.). We also acknowledge the National Institutes of Health for research support (HL 24415 to J.D.E.; DK 52356 to J.B.).
ABBREVIATIONS
- LCR
locus control region
- YAC
yeast artificial chromosome
- PFGE
pulse-field gel electrophoresis
References
- 1.Tuan D, London I M. Proc Natl Acad Sci USA. 1984;81:2718–2722. doi: 10.1073/pnas.81.9.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forrester W C, Takegawa S, Papayannopoulou T, Stammatoyannopoulos G, Groudine M. Nucleic Acids Res. 1987;15:10159–10177. doi: 10.1093/nar/15.24.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grosveld F, van Assendelft G B, Greaves D R, Kollias G. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 4.Bodine D, Ley T. EMBO J. 1987;6:2997–3004. doi: 10.1002/j.1460-2075.1987.tb02605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dillon N, Grosveld F. Nature (London) 1991;350:252–254. doi: 10.1038/350252a0. [DOI] [PubMed] [Google Scholar]
- 6.Kosteas T, Manifava M, Moschonas N, Anagnou N P. Blood. 1994;84:506. (abstr.). [Google Scholar]
- 7.Cunningham J M, Purucker M E, Jane S M, Safer B, Vanin E F, Ney P A, Lowrey C H, Nienhuis A W. Blood. 1994;84:1298–1308. [PubMed] [Google Scholar]
- 8.Stamatoyannopoulos J A, Clegg C, Li Q. Mol Cell Biol. 1997;17:240–247. doi: 10.1128/mcb.17.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langdon S D, Kaufman R E. Blood. 1998;91:309–318. [PubMed] [Google Scholar]
- 10.Bungert J, Dave U, Lieuw K E, Lim K-C, Shavit J A, Liu Q, Engel J D. Genes Dev. 1995;9:3083–3096. doi: 10.1101/gad.9.24.3083. [DOI] [PubMed] [Google Scholar]
- 11.Liu Q, Bungert J, Engel J D. Proc Natl Acad Sci USA. 1997;94:169–174. doi: 10.1073/pnas.94.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo. Cold Spring Harbor, NY: CSH Press; 1994. [Google Scholar]
- 13.Gaensler K M L, Burmeister M, Brownstein B H, Taillon-Miller P, Myers R M. Genomics. 1991;10:976–984. doi: 10.1016/0888-7543(91)90188-k. [DOI] [PubMed] [Google Scholar]
- 14.Foley K P, Leonard M W, Engel J D. Trends Genet. 1993;9:380–385. doi: 10.1016/0168-9525(93)90137-7. [DOI] [PubMed] [Google Scholar]
- 15.Gaensler K M, Kitamura M, Kan Y W. Proc Natl Acad Aci USA. 1993;90:11381–11385. doi: 10.1073/pnas.90.23.11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peterson K R, Clegg C H, Huxley C, Josephson B M, Haugen H S, Furukawa T, Stamatoyannopoulos G. Proc Natl Acad Sci USA. 1993;90:7593–7597. doi: 10.1073/pnas.90.16.7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winston J F, Chumley F, Fink G R. Methods Enzymol. 1983;101:211–228. doi: 10.1016/0076-6879(83)01016-2. [DOI] [PubMed] [Google Scholar]
- 18.Groudine M, Kohwi-Shigematsu T, Gelinas R, Stamatoyannopoulos G, Papayannopoulou T. Proc Natl Acad Sci USA. 1983;80:7551–7555. doi: 10.1073/pnas.80.24.7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoeckert C J J, Cheng H. Am J Hematol. 1996;51:220–228. doi: 10.1002/(SICI)1096-8652(199603)51:3<220::AID-AJH7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 20.Li Q, Stamatoyannopoulos J A. Mol Cell Biol. 1994;14:6087–6096. doi: 10.1128/mcb.14.9.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickinson L A, Joh T, Kohwi Y, Kohwi S T. Cell. 1992;70:631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- 22.Nakagomi K, Kohwi Y, Dickinson L A, Kohwi S T. Mol Cell Biol. 1994;14:1852–1860. doi: 10.1128/mcb.14.3.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli U K, Kas E, Poljak L, Adachi Y. Curr Opin Genet Dev. 1992;2:275–285. doi: 10.1016/s0959-437x(05)80285-0. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Z, Lin C, Wang S, Gaensler K. Blood. 1997;90:129. (abstr.). [PubMed] [Google Scholar]
- 25.Strouboulis J, Dillon N, Grosveld F. Genes Dev. 1992;6:1857–1864. doi: 10.1101/gad.6.10.1857. [DOI] [PubMed] [Google Scholar]
- 26.Stamatoyannopoulos G, Josephson B, Zhang J W, Li Q. Mol Cell Biol. 1993;13:7636–7644. doi: 10.1128/mcb.13.12.7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slightom J L, Biechl A E, Smithies O. Cell. 1980;21:627–638. doi: 10.1016/0092-8674(80)90426-2. [DOI] [PubMed] [Google Scholar]