Abstract
The bacterial leucine-responsive regulatory protein (Lrp) is a global transcriptional regulator that controls the expression of many genes during starvation and the transition to stationary phase. The Mycobacterium tuberculosis gene Rv3291c encodes a 150-amino acid protein (designated here as Mtb LrpA) with homology with Escherichia coli Lrp. The crystal structure of the native form of Mtb LrpA was solved at 2.1 Å. The Mtb LrpA structure shows an N-terminal DNA-binding domain with a helix-turn-helix (HTH) motif, and a C-terminal regulatory domain. In comparison to the complex of E. coli AsnC with asparagine, the effector-binding pocket (including loop 100–106) in LrpA appears to be largely preserved, with hydrophobic substitutions consistent with its specificity for leucine. The effector-binding pocket is formed at the interface between adjacent dimers, with an opening to the core of the octamer as in AsnC, and an additional substrate-access channel opening to the outer surface of the octamer. Using electrophoretic mobility shift assays, purified Mtb LrpA protein was shown to form a protein–DNA complex with the lat promoter, demonstrating that the lat operon is a direct target of LrpA. Using computational analysis, a putative motif is identified in this region that is also present upstream of other operons differentially regulated under starvation. This study provides insights into the potential role of LrpA as a global regulator in the transition of M. tuberculosis to a persistent state.
Keywords: LrpA, HTH motif, lat gene, EMSA, DNA binding, transcriptional regulator
Mycobacterium tuberculosis LrpA (Rv3291c) is a leucine-responsive member of the Lrp/AsnC family of transcription factors, which has representatives in a broad range of organisms spanning Eubacteria and Archaea. Members of this family have been identified in 45% of bacterial genomes and 95% of Archaea, with some species such as Mesorhizobium loti containing over a dozen paralogs (Brinkman et al. 2003). The Lrp/AsnC family has been associated with global regulation of metabolism, especially in response to changes in intracellular levels of amino acids (Calvo and Matthews 1994). They are also referred to as feast/famine regulatory proteins (FFRPs), because of their potential role in detecting and adapting to starvation conditions (Suzuki 2003). Escherichia coli contains three members of this family: LrpA, to which Mtb LrpA has 40% amino acid identity, YbaO, and AsnC, which regulate asparagine biosynthesis (Kolling and Lother 1985). Other members of this family include LrpB in Sulfolobus (Peeters et al. 2004), LrpC in Bacillus subtilis (Thaw et al. 2006), LysM in Sulfolobus (senses lysine) (Brinkman et al. 2002), Ptr2 in Methanococcus (Ouhammouch and Geiduschek 2005), and FL11 in Pyrococcus (senses glutamine) (Koike et al. 2004).
In E. coli, LrpA has been shown to influence the expression of up to 10% of the proteins in the genome, causing upregulation of some genes and downregulation of others during late-log phase (Tani et al. 2002). Leucine modulates the regulatory effects of LrpA in multiple ways. The presence of leucine potentiates some changes, antagonizes others, and has no effect for still others (Calvo and Matthews 1994). LrpA has been shown to bind directly to genomic regions upstream of a number of regulated operons, including ilvIH (Wang and Calvo 1993), leuABCD (Calvo and Matthews 1994), pap (Nou et al. 1995), and dadAX (Mathew et al. 1996). In some cases, LrpA acts as an activator to recruit the transcription machinery (e.g., ilvIH), and other times as a repressor by blocking transcription (e.g., dadAX). However, for many other genes showing differential expression, it is not known whether they are controlled directly by LrpA, or regulated indirectly through secondary transcription factors.
LrpA is strongly up-regulated during starvation (Betts et al. 2002), implicating it in response to nutrient limitation in M. tuberculosis, and therefore suggesting it might play a role in persistence (Gomez and McKinney 2004). There are several in vitro models of persistence, including stationary-phase growth (DeMaio et al. 1996; Voskuil et al. 2004) and hypoxia (Wayne and Hayes 1996; Wayne and Sohaskey 2001). Nutrient starvation is another important model for M. tuberculosis dormancy, as a nutrient-limiting environment is suspected to develop in the granulomatous lesions that encase the sites of infection (Loebel et al. 1933a,b). Betts et al. (2002) developed an in vitro system to characterize the effects of starvation on M. tuberculosis gene expression. This culture system generates apparently nonreplicating bacilli that exhibit a low level of respiration and exhibit no loss of viability in this nutrient-deprived condition for up to 6 wk. The bacilli develop resistance to isoniazid and rifampin similar to persistence in vivo (Gomez and McKinney 2004), but do not acquire susceptibility to metronidazole as in hypoxic conditions (Wayne and Sohaskey 2001), suggesting differences in the physiological responses.
In the Betts et al. (2002) starvation model, many genes appear to be down-regulated, including those that are involved in energy metabolism, lipid biosynthesis, cell division, and transcription. In contrast, starvation induces a number of stress response and virulence factors that may be important for adaptation to the host environment (Betts et al. 2002). In this model, LrpA is one of the most strongly up-regulated genes (14.6-fold after 96 h), implicating it in regulation of starvation response in Mtb. Furthermore, mutations in Lrp have been shown to provide a growth-advantage-in-stationary-phase phenotype in E. coli (Zinser and Kolter 1999). However, LrpA is only weakly up-regulated (2.9-fold) in anaerobic conditions (specifically, NRP2) (Muttucumaru et al. 2004). Deciphering the regulatory control exerted by LrpA in response to nutrient limitation will be important for understanding the nature of persistence, and ultimately for drug design.
Crystal structures for five proteins in the Lrp/AsnC family have been solved to date: LrpA in Pyrococcus furiosus (Leonard et al. 2001), FL11 in Pyrococcus OT3 (Koike et al. 2004), LrpA in E. coli (de los Rios and Perona 2006), LrpC in B. subtilis (Thaw et al. 2006), and AsnC in E. coli (Thaw et al. 2006). These structures reveal that proteins in this family typically have two domains: a classical helix-turn-helix DNA-binding domain, and a regulatory domain for binding the effector. This second domain is termed a RAM domain (regulator of amino acid metabolism) (Ettema et al. 2002), and is related to ACT domains which bind a broader range of small metabolites (Grant 2006). Lrp proteins naturally form dimers and likely interact with helical DNA as such. However, they have been reported to form a number of higher order oligomers (tetramers, hexamers, octamers), which is believed to be related to formation of histone-like particles with the DNA helix wrapped around it and making interactions at multiple evenly spaced sites (Koike et al. 2004).
In this paper, we report the crystal structure of Mtb LrpA. Mtb LrpA crystallizes as an octamer and has a similar fold to that observed previously for other members of the Lrp/AsnC family, including a helix-turn-helix DNA-binding domain and a RAM regulatory domain for binding the effector. Although largely preserved, we note several differences in the probable effector-binding pocket buried between adjacent dimers in the octamer. In addition, we identify a putative DNA binding-site motif for LrpA, in order to begin to better understand its network of control via the genes it directly regulates. The third most strongly up-regulated gene in the Betts et al. (2002) study is lat (Rv3290c), which is up-regulated 42-fold after 96 h of starvation. Lat is a lysine aminotransferase that, in other bacteria, is involved in production of antibiotics, such as cephalosporins in Streptomyces (Malmberg et al. 1993). We show by a gel-shift assay that LrpA binds in a specific fashion to a region within 150 bp upstream of the translational start site of lat. We observe a region located at −55 to −33 that resembles the AT-rich E. coli LrpA binding site pattern. We used computational methods (Gibbs sampling) to search for similar patterns upstream of other genes differentially regulated under starvation, in order to define a motif that could represent a binding site for LrpA. Interestingly, nine out of 15 instances of this motif are proximal to peaks in the curvature of the DNA, suggesting that geometry may be as important as sequence in determining the protein–DNA interaction, which has previously been reported for LrpA (Wang et al. 1994) and other family members like LrpC (Beloin et al. 2000). Among the proteins predicted to contain a motif upstream are other regulators associated with stress response, including transcription factors like whiB2, and sigma factors like sigD, supporting the hypothesis that LrpA could be acting hierarchically through a layer of secondary regulators.
Results and Discussion
The Mycobacterium tuberculosis LrpA structure
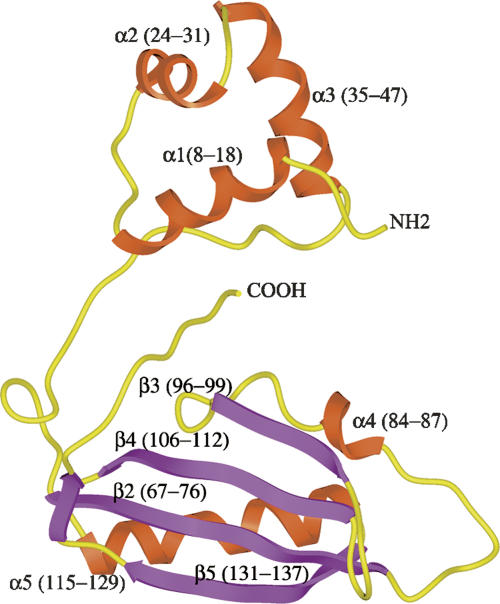
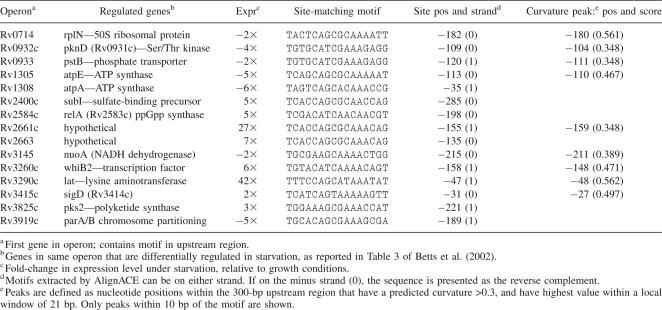
The structure of Mtb LrpA was solved by MAD methods (Hendrickson and Ogata 1997), using crystals of selenomethionylated protein formed in the orthorhombic space group C2221 (see Tables 1 and 2). Since there are no methionine residues in the native Mtb LrpA sequence (other than at the N terminus), Leu 108 was mutated to Met to obtain a selenium signal. The L108M mutation was chosen since it is a conservative substitution, and the residue in the corresponding position in the Pyrococcus FL11 sequence is also Met. The structure was refined to 2.1 Å resolution, with a final R-factor and R-free of 21% and 26%, respectively. The crystal structure of the Mtb LrpA subunit (Fig. 1) reveals an N- and C-terminal domain similar to what was observed in the structure of the Archaeal LrpA from P. furiosus (Leonard et al. 2001) and the feast/famine regulatory protein FL11 (Koike et al. 2004). The N-terminal domain contains a helix-turn-helix (HTH) motif, which has been proposed to be involved in DNA binding (Platko and Calvo 1993; Enoru-Eta et al. 2000). The α1 and α2 helices are joined by a short turn that contains a critically conserved glycine at position 32. All HTH-containing proteins characterized to date have a glycine at the analogous position in the HTH motif (Brennan and Matthews 1989). This glycine residue is thought to play an important role in forming the constrained turn of the HTH motif (Mondragon et al. 1989). The structural alignment also indicates that the HTH motif of Mtb LrpA is similar to the HTH motifs of other transcriptional regulators. Superposition of N-terminal domains of Mtb LrpA (Cα carbon atoms of residues 5–47) and the transcriptional regulator Rob (PDB: 1D5Y, Cα carbon atoms of residues 45–86) (Kwon et al. 2000), as well as the metallothionein repressor of cyanobacteria (PDB: 1SMT, Cα carbon atoms of residues 3–49) (Cook et al. 1998), have an RMSD of 2.50 Å and 2.36 Å, respectively (Fig. 2). Superposition of the E. coli Rob-DNA binary complex with Mtb LrpA showed four important residues in the α3 helix of the HTH motif of Rob (i.e., Trp 36, Gln 39, Arg 40, and Lys 43), which bond through van der Waals interactions and hydrogen bonding with the DNA major groove. Structural alignment of the HTH motif of Mtb LrpA with Rob demonstrates that Ser 36, Gln 39, Arg 41, and Arg 43 occupy analogous positions in Mtb LrpA to the above four residues in the α3 helix of Rob. A model of an Mtb LrpA–DNA complex based on the structure of Rob showed that the conserved serine 36 is close enough (2.7 Å) to form a hydrogen bond with the nitrogen of guanine 7 (DNA). In addition, the guanidino groups of Arg 43 (equivalent to Lys 43 in Rob) and Arg 41 (conserved among Lrp homologs, but not in Rob) are positioned in close proximity to, and able to make electrostatic interactions with, phosphate groups in the DNA backbone on opposite sides of the major groove. These observations suggest that residues in this helix of LrpA could determine the specificity of interaction with its cognate DNA.
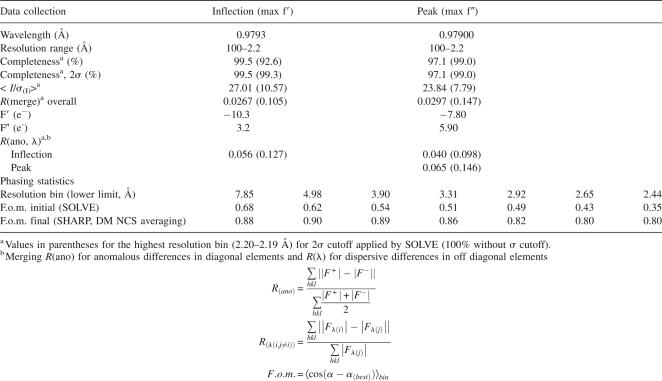
Table 1.
Anomalous data collection and phasing statistics for native LrpA structure
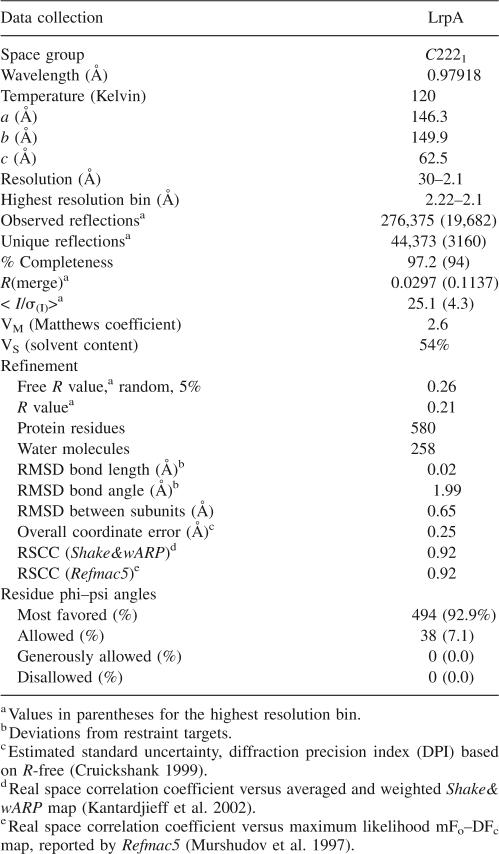
Table 2.
Data collection, refinement, and geometry statistics for native LrpA
Figure 1.
Overview of M. tuberculosis LrpA structure. Ribbon diagram of the M. tuberculosis LrpA subunit secondary structure. The helix-turn-helix (HTH) motif is formed by residues Leu 5–Ser 47. The C-terminal domain consists of residues Leu 67–Ile 137. The HTH domain and the C-terminal domain are connected by a two-hinge region, which encompasses residues Arg 48–Leu 66 and Ile 138–Pro 150. The α-helices are colored in rust, the β-strand is colored in purple, and the connecting loops are colored in yellow.
Figure 2.
Superposition of the HTH motif of M. tuberculosis LrpA (yellow) with two other transcriptional regulators, Rob of E. coli (PDB 1D5Y, purple), and the cyanobacterial metallothionein repressor (PDB 1SMT, rust).
The C-terminal domain of Mtb LrpA (i.e., residues 67–137) contains an αβ-sandwich fold similar to that of P. furiosus LrpA (Leonard et al. 2001) and the feast/famine regulatory protein FL11 (Koike et al. 2004). This region is composed of a mixed antiparallel β-sheet flanked by two α-helices (α4 and α5). This fold is commonly found in enzymes involved in amino acid metabolism and purine biosynthesis (Ettema et al. 2002). In the P. furiosus LrpA structure, the N- and C-terminal domains are connected by a single linker region (i.e., residues 50–60). In contrast, in Mtb LrpA structure the N- and C-terminal domains are connected by two flexible linker regions (i.e., residues from 44–66 and 138–150) (Fig. 1). The latter linker region (i.e., residues 138–150) in Mtb LrpA contains seven additional residues, which further extend the C-terminal end toward the N-terminal domain. As a result, the C-terminal residue, Pro 150, of Mtb LrpA extends far toward the N-terminal domain, forming a favorable position to interact with the N-terminal Arg 14 residue, which could further stabilize the monomer.
The rotation angle between the N- and C-terminal domains varies by up to 20 degrees among different members of the Lrp/AsnC family (de los Rios and Perona 2006). The relative orientation of the two domains in Mtb LrpA most closely resembles that in the E. coli AsnC structure (Thaw et al. 2006).
Mtb LrpA oligomerization
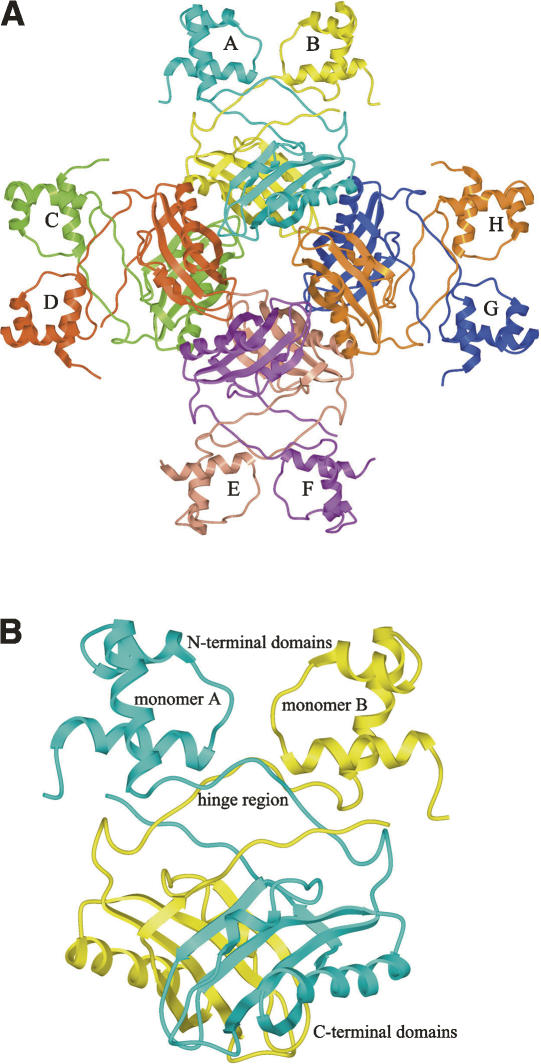
Both Mtb LrpA and P. furiosus LrpA appear to exist as dimers, tetramers, and octamers in solution (Leonard et al. 2001; Shrivastava et al. 2004). In the crystal lattice, the asymmetric unit contains four molecules of Mtb LrpA. The intersubunit interactions are responsible for the formation of the quaternary assembly (octamer). It consists of four tightly packed dimers arranged in D4 symmetry (Fig. 3A), similar to all the other Lrp structures except E. coli LrpA, in which the octamer appears partially “open” when crystallized in the presence of DNA (de los Rios and Perona 2006). The solvent-accessible surface area for an individual Mtb LrpA subunit is 10,481 Å. Dimerization of the subunits buries 2643 Å of each subunit's surface area, which represents approximately one-quarter of the total subunit surface area. The subunit–subunit interactions of the dimer appear to be strong (Fig. 3B), as there are 22 hydrogen bonds between the subunits at the dimer interface. This hydrogen bonding, as well as the significant van der Waals interactions that occur between the two subunits, primarily involve residues within the four antiparallel β-strands of the C-terminal domain. Specifically, residues from the β2 strand of one subunit form eight hydrogen bonds with residues from the β4 strand, and residues from the β5 strand form four hydrogen bonds with the C-terminal hinge region of the neighboring subunit. Additional subunit interactions occur between the linker regions (i.e., residues 50–59) and the α1-helix.
Figure 3.
(A) Schematic representation of the M. tuberculosis LrpA octamer, showing the dimer–dimer interactions that lead to the formation of a stable octamer. Each subunit is labeled A–H, and colored as follows: A, cyan; B, yellow; C, light green; D, rust; E, light salmon; F, purple; G, blue; H, orange. (B) Schematic representation of the M. tuberculosis LrpA dimer, with local twofold noncrystallographic symmetry, demonstrating the LrpA subunit–subunit interactions that promote the formation of a stable dimer. Subunit A is colored in cyan and subunit B is colored in yellow.
Oligomerization of the dimers into an octamer buries 571 Å per monomer, which is only 5.4% of each monomer surface area. As with the dimer formation the octamer interface is formed primarily from residues in the C-terminal domain. Hydrogen bonding is not nearly as extensive (12 hydrogen bonds) at the octamer interface as it is with the dimer interface (22 hydrogen bonds).
Comparison of the structure of Mtb LrpA with structures of homologs
A structural similarity search of the Mtb LrpA subunit using DALI (Holm and Sander 1995) revealed high similarity (14.4 Z-Score) to P. furiosus LrpA (PDB code 1I1G). Vector alignment search tool (VAST) (Gibrat et al. 1996) revealed high similarity with the Archeal feast/famine regulatory protein, FL11 (PDB code 1RI7; VAST score 10.9) and structural similarity with P. furiosus LrpA (VAST score 10.7). The Mtb LrpA structure (140 Cα atoms) superimposed very well with P. furiosus LrpA (140 Cα atoms). Sequence alignment shows that Mtb LrpA has ∼23% sequence identity with the feast/famine regulatory protein FL11 (Fig. 4). Despite the relatively low level of amino acid sequence identity between Mtb LrpA and the feast/famine regulatory proteins, least-square superposition of the 145 Cα carbon atoms indicates close resemblance (RMSD of 2.6 Å).
Figure 4.
Sequence alignment of Lrp homologs from E. coli, P. furiosus, M. tuberculosis, and AsnC. The top line asterisk indicates amino acids that can potentially interact with DNA. Multiple alignment was generated with Clustal W 1.8.2 (Thompson et al. 1994). Perfectly conserved residues are shown in boxes.
A DALI search further revealed that the Mtb LrpA C-terminal domain has significant similarities to the Archaeoglobus fulgidus (PDB 1P9Q) RAM domain (Z = 7.1), the human elongation factor (PDB 1B64, Z = 6.5), and the Streptococcus aureus nickel-responsive repressor (PDB 1OKR, Z = 7.0). In addition, the C-terminal domain (Cα carbon atoms of residues from 67–137) of Mtb LrpA and P. furiosus LrpA shows significant resemblance to the ACT (Aspartokinase, Chorismate mutase, and TyrA) domain (Cα carbon atoms of residues from 335–410) of E. coli SerA (Grant et al. 1996).
The ACT domain folds as an αβ sandwich with ferredoxin-like βαββαβ topology (Grant et al. 1996; Al-Rabiee et al. 1996). The effects of ligand binding on proteins with ACT-like regulatory domains vary. For example, phenylalanine hydroxylase is activated by a regulatory ligand (Chipman and Shaanan 2001), whereas tyrosine binding to TyrR protein represses transcription of a group of operons. In E. coli D-3-phosphoglycerate dehydrogenase (PGDH), the ACT domain binds to L-serine, which is the final product of the serine biosynthetic pathway and also acts as an allosteric regulator of PGDH (Grant et al. 1996). Superposition of the Mtb LrpA C-terminal RAM domain (Cα carbon atoms of residues from 67–137) over the ACT domain of E. coli PGDH (Cα carbon atoms of residues from 335–410), reveals close structural resemblance (RMSD 2.48 Å). However, the dimer interactions in these two proteins differ significantly from one another. In PGDH, the interaction between the two ACT domains occurs through the α2 helix and the β3 strand, whereas Mtb LrpA dimerization occurs through interactions of antiparallel β-sheets that face each other. In the ACT domain of E. coli PGDH, the effector-binding residues are located at the dimer interface within the α1 and β1 strands and are highly conserved. In contrast, in the Mtb LrpA structure, residues in the corresponding strands are poorly conserved, suggesting that the effector-binding sites in the two proteins are very different.
Contact between adjacent dimers appears to involve two sets of interactions during oligomerization; residues from the connecting loops between the β2 strand and α4 helix (i.e., Pro 78, Ser 79, and Gln 80) interact through hydrogen bonds with the α5 helix (i.e., Gln 123, Arg 132, and Thr 127) of a noncrystallographic symmetry-related subunit. Similarly, residues from the loop connecting the β3 and β4 strands of one subunit (e.g., Gly 102) interact with backbone atoms of the β5 strand of the other subunit. Hence, the C-terminal domain is important for dimerization, as well as oligomerization of dimers into higher order oligomers, such as tetramers, octamers, and hexadecamers, as previously observed for E. coli Lrp (Chen and Calvo 2002). Mutational studies have shown that seven mutations in the C-terminal domain of E. coli Lrp result in the loss of sensitivity to leucine (Platko and Calvo 1993). When the locations of these mutations were mapped on the Mtb LrpA structure, six of the seven were found to be located at the interfaces between subunits (Ser 99, Ser 105, Ala 115, Ile 137, Ile 138, and Leu 139).
Based on the crystal structure of the E. coli AsnC structure in complex with asparagine, the effector-binding site was identified to be in a surface pocket formed by two subunits at the interface between adjacent dimers (Thaw et al. 2006). The pocket in LrpA is formed primarily from loop 99–106 in one subunit and residues from strand β5 and helix α5 of another subunit. The Mtb LrpA structure also shows a cavity between dimers in the molecular surface of the octamer assembly, with a substrate-access channel opening into the core of the octamer near Glu 103, as in the AsnC structure. However, there is an additional access channel to the pocket from an opening to the outer surface of the complex near Glu 119 (which is blocked off by Tyr 82 in the AsnC–asparagine complex, and by Phe 90 in the E. coli LrpA structure). Based on a homology model of E. coli LrpA based on the AsnC structure, Thaw et al. (2006) predicted the leucine-binding pocket of LrpA to consist of residues Phe 90, Leu 108, Val 109, Gly 111, Asp 114, Leu 136, Thr 144, and Thr 146. In the Mtb LrpA structure, the residues that actually line this pocket include Ser 99a–Ser 105a (primarily backbone atoms), Tyr 106a, Leu 118b, Glu 119b, Leu 122b, Arg 126, Thr 133b, and Ser 135b (where a and b represent different subunits). The GD/E motif in the center of the loop (residues 102–103 in LrpA) is highly conserved among Bacteria, as well as Archaea; several carbonyl oxygens in this loop make hydrogen bonds with the main-chain atoms of the bound asparagine in AsnC, are structurally conserved in Mtb LrpA, and would be consistent with binding leucine in a similar fashion. However, there are significant differences where the side chain resides. In AsnC, the carboxamide of the asparagine ligand hydrogen bonds with the backbone carbonyl of Ser 105 and carboxamide of Gln 128 (2.7 Å each). In LrpA, Ser 105 is conserved, and Arg 126 occupies approximately the same position in the pocket as Gln 128 in AsnC. It is notable that Ser 105 is one of two leucine-response mutants identified in E. coli LrpA by Platko and Calvo (1993) that can be mapped onto the central loop in this pocket (Ser 99 and Ser 105 correspond to Leu 108 and Asp 114 in E. coli LrpA, respectively). In addition, the aliphatic part of Glu 119b and Leu 139c (from a third subunit) contribute to the hydrophobicity of the pocket. These observations support the hypothesis that leucine could bind LrpA in the same location as asparagine in AsnC.
LrpA binds to the lat promoter
Among the genes that are up-regulated under starvation (Betts et al. 2002), three are located immediately downstream of lrpA. These include lat (Rv3290c), Rv3289c, and usfY (Rv3288c), which are also among the most highly induced M. tuberculosis genes during nutrient starvation (Betts et al. 2002). Hence, the lat operon was used as a representative potential target of LrpA regulation in this study. The lat gene encodes lysine ε-aminotransferase, which converts L-lysine into L-pipecolic acid by transferring the ε-amino group of L-lysine to α-ketoglutarate, and is considered to be the rate-limiting step in the biosynthesis of β-lactam antibiotics (Malmberg et al. 1993). Rv3289c is annotated as a possible transmembrane protein, and the function of UsfY is also currently unknown (http://genolist.pasteur.fr/TubercuList/).
Electrophoretic mobility shift assays (EMSA) were used to determine the specific interaction of Mtb LrpA with the lat upstream sequences. When the extended upstream lat sequence (−300 to +1 region) was incubated with purified M. tuberculosis LrpA, a single protein–DNA complex was observed (Fig. 5A). In contrast, no protein–DNA complexes were observed when the −300 to −150 region upstream of lat was used in the EMSA (data not shown), suggesting that M. tuberculosis LrpA binds more proximally to the upstream region of lat. We then examined the −150 to +1 region upstream of lat to identify a potential core sequence for M. tuberculosis LrpA binding and found a 23-bp sequence, CATAATTTTTCCAGCATAAATAT (−55 to −33), that resembles the AT-rich consensus sequence for Lrp binding in E. coli: YAGHAWATTWTDCTR, where Y = C/T, H = not G, W = A/T, D = not C, and R = A/G (Cui et al. 1995).
Figure 5.
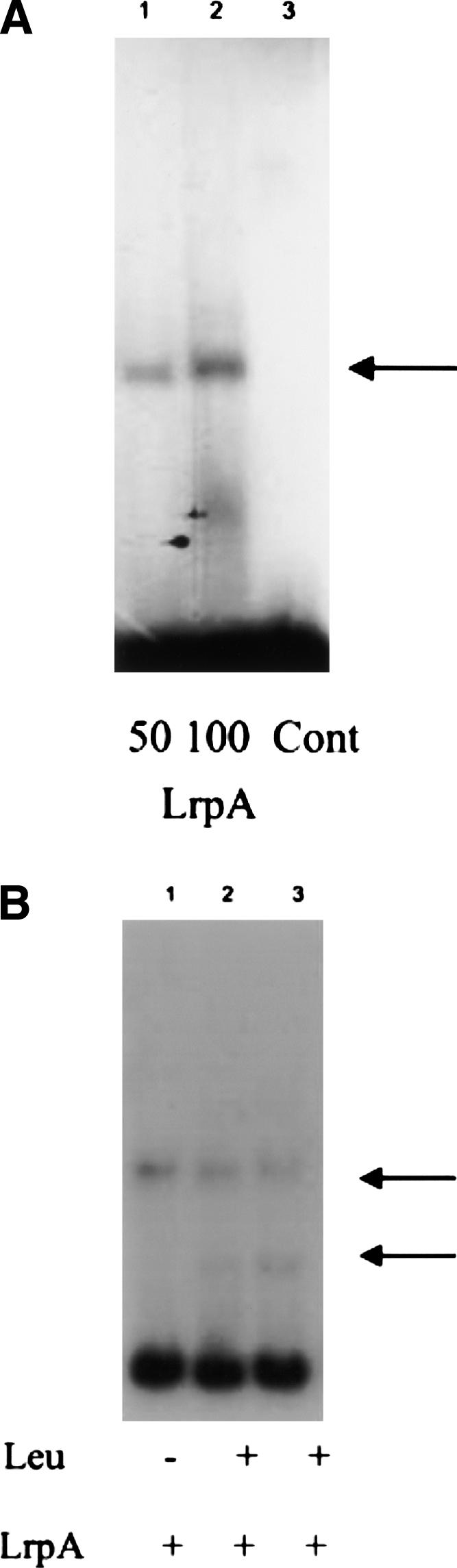
Binding of LrpA to the lat upstream region shown by the electrophoretic mobility shift assay (EMSA). (A) Gel shift was performed with (15 fmol) 300-bp fragment (−300 to +1) encompassing the promoter and transcription initiation site of the lat gene. Lanes 1 and 2: 50 and 100 ng of LrpA, respectively; lane 3: without LrpA (control). (B) The assay was performed with 100 ng of purified M. tuberculosis LrpA and the 23-bp fragment (15 fmol) in the presence and absence of 10 mM or 20 mM leucine. Lane 1: 100 ng of LrpA with no leucine; lane 2: 100 ng of LrpA with 10 mM leucine; lane 3: 100 ng LrpA with 20 mM leucine.
To determine the effect of leucine on the binding of Mtb LrpA to the lat promoter, an EMSA (gel-shift assay) was performed using Mtb LrpA and two different concentrations of L-leucine (10 mM and 20 mM). The binding of purified recombinant M. tuberculosis LrpA to the 23-bp fragment resulted in a single protein–DNA complex in the absence of leucine (Fig. 5B). On the other hand, we observed an additional faster migrating complex and a relative decline in the intensity of the original slower complex in the presence of 10 mM leucine (Fig. 5B). Moreover, the addition of 20 mM leucine increased the intensity of the second faster complex and further decreased the intensity of the slower complex (Fig. 5B). These findings suggest that the addition of leucine alters the binding of M. tuberculosis LrpA to the lat upstream sequences, in a manner similar to that described for leucine interaction with the E. coli Lrp at certain target sites (Calvo and Matthews 1994). The faster migrating complex induced by the addition of leucine could reflect partial dissociation, e.g., of hexadecamers into octamers, as suggested by Chen and Calvo (2002).
Computational identification of putative LrpA binding sites
Given the results of the gel-shift assays that suggested that LrpA binds to a genomic region upstream of lat, we used Gibbs sampling to identify putative regulatory motifs in this region that are shared by other genes differentially regulated during starvation, and used these motifs to search the rest of the genome to identify other genes potentially regulated by LrpA. As a representative list of genes whose expression is influenced by starvation, we took the 114 genes listed in Table 3 of Betts et al. (2002), whose expression level changed significantly 96 h after transfer to PBS, (phosphate-buffered saline) including both up- and down-regulated genes. All four genes in the lat operon—Rv3291c (lrpA), Rv3290c (lat), Rv3289, and Rv3288 (usfY)—were observed to be strongly up-regulated (10- to 40-fold) and were included in this list.
Table 3.
Regulated genes containing sites in upstream regulatory regions matching the putative LrpA DNA-binding motif identified by AlignACE
Next, the genes were mapped to their operons, since regulatory motifs would be expected to occur upstream of the first gene in each operon. The operon definitions were obtained from http://www.webtb.org, which were derived based on a combination of proximity, experimental evidence, and other information (Moreno-Hagelsieb and Collado-Vides 2002). After removing redundancies, this mapping left 68 genes representing operons containing genes whose expression level were altered during starvation. A genomic region consisting of 300 bp upstream of the translational start site of each of these genes was extracted (taking the appropriate reverse complement for genes on the minus strand of the chromosome). The region upstream of Rv3290c (lat) was also inlcuded, even though it is predicted to be the second gene in the operon headed by Rv3291c (lrpA), since there is direct evidence the LrpA binds upstream of it. Despite the fact that operon-prediction algorithms suggest lrpA and lat are the first two genes in a common operon, this does not preclude the possibility that they could be independently transcribed. In fact, there is precedent for this phenomenon, as dosR, a transcriptional regulator involved in the hypoxic response, appears to have its own independent promoter, separate from the promoter upstream of the Rv3134c-dosR-dosS operon in which it resides (Bagchi et al. 2005).
These 68 DNA sequences were subjected to motif analysis using AlignACE (Neuwald et al. 1995), which uses Gibbs sampling to identify short (10–20-bp) fragments that are statistically overrepresented in the set. The patterns do not have to occur in every sequence, can occur more than once in a given sequence, are searched in both orientations, and are not positionally constrained (can occur in any position in any sequence—not required to be aligned). The estimation of statistical significance is strongly biased by assumptions about background nucleotide distribution, so the option “−gcback 0.65” was used to reflect the high GC-content of the Mtb genome. A total of 85 motifs were identified, with MAP scores ranging from 29.7 to 0.08.
To evaluate the relevance of these motifs, ScanACE (Hughes et al. 2000) was used to search the upstream regions (300 bp) of all the genes in the H37Rv genome and calculate a specificity score, which is a statistic that indicates the degree of overlap between the genes with a hit (containing a motif upstream) and the original set of 68 genes representing regulated operons. Better (lower) scores are achieved when a motif occurs in more of the regulated genes and/or fewer genes in the rest of the genome. Specificity scores ranged between 0.1 and 3e-16. Motif 46 had a MAP score of 7.9 and a specificity of 3.2e-11. Instances of it occurred in 15 genes from the regulated set (using a threshold set so that no more than ∼100 false positive hits are found in the rest of the genome upstream of unregulated genes). Importantly, the upstream region of lat (Rv3290c) contained this motif. In fact, the motif appears at position −48 to −33 (TTTCCAGCATAAATAT), within the 150-bp region identified by the gel-shift studies and coincident with the AT-rich site (−55 to −33) that bears resemblance to the E. coli Lrp binding pattern (Cui et al. 1995). The motif occurred at varying positions and in both orientations in the upstream regions of the other genes, from −22 to −298, without any apparent consistency. The actual pattern is a probabilistic profile generalized over the individual instances of the motif found, and is illustrated by the Logo plot shown in Figure 6. The other genes showing differential expression under starvation that contain instances of this motif are listed in Table 3.
Figure 6.
Logo plot of LrpA DNA-binding motif identified by AlignACE (generated using http://weblogo.berkeley.edu).
As partial validation of this motif, we analyzed the proximity of the hits to predicted peaks in the local curvature of the DNA. Wang et al. (1994) showed that LrpA in E. coli preferentially binds at locations upstream of the ilvIH operon showing intrinsic curvature. It has been reported that LrpC, another member of the AsnC/LrpA transcription-factor family in B. subtilis, binds in a nonsequence-specific fashion to its own upstream region, and its binding is also centered on a region that shows a bend experimentally (Beloin et al. 2000; Tapias et al. 2000). Bends in DNA have also been associated with other bacterial transcription factor binding sites (Asayama and Ohyama 2005), and may be affected by supercoiling (Beloin et al. 2003), which could be involved in stress response and/or transition to stationary phase.
To evaluate bending in the upstream regions near LrpA binding sites, we used the program dnacurve.py (http://www.lfd.uci.edu/∼gohlke/code/dnacurve.py.html) to predict the curvature of DNA based on the dinucleotide wedge model (Goodsell and Dickerson 1994) (twist/roll/tilt parameters: “aawedge,” window size: 21), and looked for proximity of motifs to predicted peaks in the curvature in the upstream regions. Of the 15 genes containing the motif (identified in Table 3), nine genes were found to have a peak in the curvature with a magnitude >0.3 within 10 bp up- or downstream of the predicted site. For comparison, there are on average ∼2.2 such peaks upstream of each of the 3989 in the TB genome. The probability of a motif in an arbitrary position within these 300 bp regions being within 10 bp of such a peak is 16.5%. Using the binomial distribution, the probability that nine out of 15 genes contain motifs within 10 bp of a peak in the curvature is 0.0002, suggesting that this association is highly nonrandom. The proximity of a motif to a peak in the curvature was not systematically associated with up- or down-regulation.
There are several notable genes in this set that can be rationalized in terms of regulation of gene expression under starvation. Genes such as pstB (phosphate transporter), atpA/E (ATPase), subI (sulfate binding), nuoD (NADH dehydrogenase), and pks2 (polyketide synthase) could be involved in response to changes in environment and an attempt to import or synthesize new metabolites at the onset of starvation. whiB2 is a transcription factor in the WhiB family, associated with sporulation in Streptomyces. whiB2, one of seven members of this family in the Mtb genome, is specifically down-regulated in the late-stationary phase (Geiman et al. 2006). It is possible that Lrp is controlling whiB2 as a secondary regulator in a regulatory cascade, allowing Lrp to exert a more global influence over expression indirectly through genes like whiB2, even if it does not bind directly to regions upstream of differentially regulated genes itself.
A connection between RelA and Lrp has previously been noted (Landgraf et al. 1996; Tani et al. 2002). RelA synthesizes ppGpp (guanosine tetraphosphate) at ribosomes when they encounter uncharged tRNAs, and thus helps to signal growth-limiting conditions, as part of the so-called “stringent response” (Dahl et al. 2003). Furthermore, ppGpp itself can up-regulate Lrp expression (Landgraf et al. 1996). It would make sense for Lrp to up-regulate RelA contemporaneously for this signaling interaction to be effective. Finally, Lrp appears to have a binding site in the region upstream of sigD. sigD is a sigma factor for a subset of genes with a unique promoter (Calamita et al. 2005), and many of these genes are associated with starvation response (particularly, heat/cold-shock proteins, lipid metabolism, and cell-wall biosynthesis enzymes, and various transcription factors) (Raman et al. 2004). It is unknown exactly what induces sigD expression, but regulation by Lrp would help explain the broad range of effects of starvation on expression levels through this secondary control.
Materials and Methods
Cloning and expression of Mtb lrpA
A 450-bp DNA fragment containing the lrpA (Rv3291c) gene was amplified by PCR, with M. tuberculosis H37Rv genomic DNA as a template, using the following oligonucleotide primers: 5′-AGATGAAGCATATGAACGAGGCGCTCGACGAT-3′ and 5′-AGAGTAAGCTTATGGTATATGCTGCCTATCGC-3′. The amplified DNA fragment was digested with NdeI and HindIII restriction enzymes and subcloned into the corresponding restriction sites in the pET28b vector containing an N-terminal His tag (Novagen). Production of a selenomethionylated (Se-Met) protein for multiple anomalous dispersion (MAD) phasing was facilitated by mutation of Leu (TTG) at position 108 to Met (ATG), using a QuikChange site-directed mutagenesis kit (Stratagene). Se-Met protein was prepared according to published methods (Miller 1972). The pET28b-lrpA recombinant plasmid was transformed into E. coli B834 (DE3), a Met auxotroph strain. The transformed cells were grown to exponential phase at 37°C in LB medium containing kanamycin. For production of Se-Met-labeled protein, the cells were grown in M9 minimal media, supplemented with all 19 standard amino acids and selenomethionine. Expression of LrpA was then induced using 1 mM IPTG, and cells were harvested after growth for 14 h at 20°C.
Mtb LrpA purification
The harvested cells were pelleted and resuspended in buffer A (20 mM Tris-HCl, pH 7.5, 50 mM imidazole) containing 1 mM PMSF and complete EDTA-free protease inhibitors (Roche). The cells were lysed using a French press, and the cell suspension was centrifuged at 15,000g for 1 h. The clear supernatant was loaded onto a Pharmacia Hi-trap Ni2+-chelating column and washed with 300 mL buffer A containing 500 mM NaCl. The bound M. tuberculosis LrpA was eluted from the nickel affinity column using 20 mM Tris-HCl, pH 8.0, 500 mM imidazole, and 500 mM NaCl. The eluant was concentrated by Centriprep (Amicon) to 7.0 mg/mL and applied onto a Sephadex 200 gel-filtration column (Amersham Biosciences), equilibrated with 20 mM Tris-HCl, 1 mM EDTA, and (1 mM) dithiothreitol (pH 7.5). The purified protein was dialyzed in a buffer containing 20 mM Tris/HCl, pH 7.5, 50 mM NaCl, 10% glycerol, and 0.5 mM EDTA, and concentrated in a Centriprep-3 filter prior to crystallization. The protein was >95% pure, as observed by SDS-PAGE.
Crystallization
Crystallization of the apo-protein was carried out in hanging-drops, using the factorial screening method (Hampton Research). Diffraction-quality crystals were obtained for native M. tuberculosis LrpA (7 mg/mL in 20 mM Tris/HCl, pH 7.5, 50 mM NaCl, 10% glycerol, and 0.5 mM EDTA), equilibrated against 500 μL of well solution containing 70 mM sodium acetate trihydrate, pH 6.0, 30% glycerol, and 5.6% PEG 4000 as a precipitant. Crystals (0.2 × 0.3 × 0.3 mm) grew at 16°C within 2–5 d in 4 μL hanging-drops (2 μL of LrpA combined with 2 μL of well solution).
Data collection and processing
Highly redundant and complete selenium K-edge MAD diffraction data from a single Se-Met-LrpA crystal was collected at two wavelengths, using an ADSC CCD detector on beamline 14-ID at the Advanced Photon Source (APS) of the Argonne National Laboratory (ANL). Crystals mounted in cryo-loops were flash-cooled in a liquid N2 stream (120 K) after brief soaks in N-paratone oil. The diffraction data were reduced using DENZO (Otwinowski and Minor 1997), and intensities were scaled with SCALEPACK (Evans 1993). The reflections were indexed centered orthogonal (a = 146.3 Å, b = 149.9 Å, c = 62.5 Å). Examination of the integrated and scaled data indicated the orthorhombic space group C2221. Solvent content calculations (Matthews 1968) indicated the presence of a tetramer (VM 2.6, VS 54%) in the asymmetric unit.
Structure determination
Experimental phases for M. tuberculosis LrpA were obtained by MAD phasing (Hendickson and Ogata 1997) (Table 1). SHELXD located the four Se sites in the asymmetric unit, consistent with a tetramer in the asymmetric unit (Sheldrick and Gould 1995), and SOLVE (Terwilliger and Berendzen 1999) was used to refine the sites and calculate initial protein phases, resulting in an overall figure of merit of 0.41 for the data in the resolution range of 100–2.2 Å. Further phase improvement with solvent flattening in autoSHARP (Vonrhein et al. 2006) followed by DM (Cowtan and Main 1996) resulted in density-modified maps of high quality showing clear electron density for all four molecules of protein in the asymmetric unit. The electron density map was submitted to TEXTAL (Ioerger and Sacchettini 2003) for automated model building. After determining the NCS operator from the Se-substructure using graphical analysis and refinement with LSQKAB (CCP4), the electron density was averaged and solvent-flattened using DM (Cowtan and Zhang 1999). Starting from the TEXTAL tracing, all of the M. tuberculosis LrpA residues, except the initial five amino acids, could be built into the density using XTALVIEW (McRee 1999). Water molecules were manually added during iterative cycles of model building and refinement using a Fourier difference map.
Several cycles of manual model building, and NCS-restrained maximum likelihood refinement in REFMAC5, yielded a high-quality model with an R-factor of 21.0% and R-free of 26.0% (Table 2) for the native LrpA structure. The protein structure file for Mtb LrpA has been deposited in the RCSB with the PDB code 2QZ8.
Electrophoretic mobility shift assay (EMSA)
DNA binding of purified M. tuberculosis LrpA to the lat (Rv3290c) promoter region was determined by an EMSA analysis. A 300-bp segment of genomic DNA covering the extreme 5′-terminus of the lat promoter region was prepared by PCR amplification, using the forward primer 5′ GGCGTTCGTGGCTATCACTCCTCTT-3′ and reverse primer 5′-CATGACGCTATGATAGCAGGAATA-3′. The amplified product was labeled at the 3′ end by a filling reaction, using [α-P32]-dATP (specific activity, 25,000 cpm), labeling mix-ATP, and 5 units of Klenow fragment of DNA. The standard reaction (15 mL) contained 100 mM Tris-HCl (pH 7.5), 500 mM NaCl, 5 mM DTT, 50% glycerol, 100 mM MgCl2, 1 μg/mL poly (dI-dC), and 15 fmol P32-labeled probe. Purified protein of M. tuberculosis LrpA was incubated in the reaction mixture for 30 min. Following incubation, the reaction mix was applied to separate lanes of a 5% polyacrylamide gel in a buffer containing 22 mM Tris-boric acid for 2 h. The gel was removed and exposed to X-ray film for 2–6 h. Protein–DNA complexes were detected by autoradiography. After determining the binding site through PCR deletion analysis, a 23-bp DNA fragment (CATAATTTTTCCAGCATAAATAT) was generated from the complementary single-stranded sequence that was chemically synthesized and annealed. The EMSA was performed as described above, and leucine was added at different concentrations (10 mM or 20 mM) and incubated for 20 min in the reaction mix before analysis.
Acknowledgments
We acknowledge Joshua L. Owen for many helpful discussions and Prianka Desai for excellent technical assistance. We thank the scientists of beamline 14-ID at the Advanced Photon Source (APS) of the Argonne National Laboratory (ANL) for their help in data collection. This work was supported by National Institutes of Health grant PO1-AIO68135 and by the Robert A. Welch Foundation (A-0015). We would like to thank Dr. Morad Hassani from Albert Einstein College of Medicine for his thoughtful comments.
Footnotes
Reprint requests to: James C. Sacchettini, Department of Biochemistry and Biophysics, Texas A&M University, College Station, TX 77843-2128, USA; e-mail: sacchett@tamu.edu; fax: (979) 862-7638.
Abbreviations: LrpA, leucine-responsive protein; lat, lysine ε-aminotransferase; HTH, helix-turn-helix; MAD, multiwavelength anomalous dispersion; RAM, regulator of amino acid metabolism; EMSA, electrophoretic mobility shift assay; ACT domain, aspartokinase/chorismate mutase/TyrA domain; FFRPs, feast/famine regulatory proteins; PGDH, D-3-phosphoglycerate dehydrogenase.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073192208.
References
- Al-Rabiee, R., Lee, E.J., Grant, G.A. The mechanism of velocity modulated allosteric regulation in D-3-phosphoglycerate dehydrogenase. Cross-linking adjacent regulatory domains with engineered disulfides mimics effector binding. J. Biol. Chem. 1996;271:13013–13017. doi: 10.1074/jbc.271.22.13013. [DOI] [PubMed] [Google Scholar]
- Asayama, M., Ohyama, T. Curved DNA and prokaryotic promoters: A mechanism for activation of transcription. In: Ohyama T., editor. DNA conformation and transcription. Springer; Berlin, Germany: 2005. [Google Scholar]
- Bagchi, G., Chauhan, S., Sharma, D., Tyagi, J.S. Transcription and autoregulation of the Rv3134c-devR-devS operon of Mycobacterium tuberculosis . Microbiol. 2005;151:4045–4053. doi: 10.1099/mic.0.28333-0. [DOI] [PubMed] [Google Scholar]
- Beloin, C., Exley, R., Mahé, A.L., Zouine, M., Cubasch, S., Le Hégarat, F. Characterization of LrpC DNA-binding properties and regulation of Bacillus subtilis lrpC gene expression. J. Bacteriol. 2000;182:4414–4424. doi: 10.1128/jb.182.16.4414-4424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloin, C., Jeusset, J., Révet, B., Mirambaut, G., Le Hégarat, F., Le Cam, E. Contribution of DNA conformation and topology in right-handed DNA wrapping by the Bacillus subtilis LrpC protein. J. Biol. Chem. 2003;278:5333–5342. doi: 10.1074/jbc.M207489200. [DOI] [PubMed] [Google Scholar]
- Betts, J.C., Lukey, P.T., Robb, L.C., McAdam, R.A., Duncan, K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- Brennan, R.G., Matthews, B.W. The helix-turn-helix DNA binding motif. J. Biol. Chem. 1989;264:1903–1906. [PubMed] [Google Scholar]
- Brinkman, A.B., Bell, S.D., Lebbink, R.J., de Vos, W.M., van der Oost, J. The Sulfolobus solfataricus Lrp-like protein LysM regulates lysine biosynthesis in response to lysine availability. J. Biol. Chem. 2002;277:29537–29549. doi: 10.1074/jbc.M203528200. [DOI] [PubMed] [Google Scholar]
- Brinkman, A.B., Ettema, T.J., de Vos, W.M., van der Oost, J. The Lrp family of transcriptional regulators. Mol. Microbiol. 2003;48:287–294. doi: 10.1046/j.1365-2958.2003.03442.x. [DOI] [PubMed] [Google Scholar]
- Calamita, H., Ko, C., Tyagi, S., Yoshimatsu, T., Morrison, N.E., Bishai, W.R. The Mycobacterium tuberculosis SigD sigma factor controls the expression of ribosome-associated gene products in stationary phase and is required for full virulence. Cell. Microbiol. 2005;7:233–244. doi: 10.1111/j.1462-5822.2004.00454.x. [DOI] [PubMed] [Google Scholar]
- Calvo, J.M., Matthews, R.G. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli . Microbiol. Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., Calvo, J.M. Leucine-induced dissociation of Escherichia coli Lrp hexadecamers to octamers. J. Mol. Biol. 2002;318:1031–1042. doi: 10.1016/S0022-2836(02)00187-0. [DOI] [PubMed] [Google Scholar]
- Chipman, D.M., Shaanan, B. The ACT domain family. Curr. Opin. Struct. Biol. 2001;11:694–700. doi: 10.1016/s0959-440x(01)00272-x. [DOI] [PubMed] [Google Scholar]
- Cook, W.J., Kar, S.R., Taylor, K.B., Hall, L.M. Crystal structure of the cyanobacterial metallothionein repressor SmtB: A model for metalloregulatory proteins. J. Mol. Biol. 1998;275:337–346. doi: 10.1006/jmbi.1997.1443. [DOI] [PubMed] [Google Scholar]
- Cowtan, K.D., Main, P. Phase combination and cross validation in iterated density-modification calculations. Acta Crystallogr. D Biol. Crystallogr. 1996;52:43–48. doi: 10.1107/S090744499500761X. [DOI] [PubMed] [Google Scholar]
- Cowtan, K.D., Zhang, K.Y. Density modification for macromolecular phase improvement. Prog. Biophys. Mol. Biol. 1999;72:245–270. doi: 10.1016/s0079-6107(99)00008-5. [DOI] [PubMed] [Google Scholar]
- Cruickshank, D.W. Remarks about protein structure precision. Acta Crystallogr. D Biol. Crystallogr. 1999;55:583–601. doi: 10.1107/s0907444998012645. [DOI] [PubMed] [Google Scholar]
- Cui, Y., Wang, Q., Stormo, G.D., Calvo, J.M. A consensus sequence for binding of Lrp to DNA. J. Bacteriol. 1995;177:4872–4880. doi: 10.1128/jb.177.17.4872-4880.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl, J.L., Kraus, C.N., Boshoff, H.I.M., Doan, B., Foley, K., Avarbock, D., Kaplan, G., Mizraji, V., Barry, C.E. The role of Rel(Mtb)-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. PNAS. 2003;100:10026–10031. doi: 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Rios, S., Perona, J.J. Structure of the Escherichia coli leucine-responsive regulatory protein Lrp reveals a novel octameric assembly. J. Mol. Biol. 2006;366:1589–1602. doi: 10.1016/j.jmb.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMaio, J., Zhang, Y., Ko, C., Young, D.B., Bishai, W.R. A stationary-phase stress-response σ factor from Mycobacterium tuberculosis . Proc. Natl. Acad. Sci. 1996;93:2790–2794. doi: 10.1073/pnas.93.7.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoru-Eta, J., Gigot, D., Thia-Toong, T.L., Glansdorff, N., Charlier, D. Purification and characterization of Sa-Lrp, a DNA-binding protein from the extreme thermoacidophilic archaeon Sulfolobus acidocaldarius homologous to the bacterial global transcriptional regulator Lrp. J. Bacteriol. 2000;182:3661–3672. doi: 10.1128/jb.182.13.3661-3672.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettema, T.J., Brinkman, A.B., Tani, T.H., Rafferty, J.B., Van Der Oost, J. A novel ligand-binding domain involved in regulation of amino acid metabolism in prokaryotes. J. Biol. Chem. 2002;277:37464–37468. doi: 10.1074/jbc.M206063200. [DOI] [PubMed] [Google Scholar]
- Evans, S.V. SETOR: Hardware lighted three-dimensional solid model representation of macromolecules. J. Mol. Graph. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- Geiman, D.E., Raghunand, T.R., Agarwal, N., Bishai, W.R. Differential gene expression in response to exposure to antimycobacterial agents and other stress conditions among seven Mycobacterium tuberculosis whiB-like genes. Antimicrob. Agents Chemother. 2006;50:2836–2841. doi: 10.1128/AAC.00295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibrat, J.F., Madej, T., Bryant, S.H. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- Gomez, J.E., McKinney, J.D. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis (Edinb.) 2004;84:29–44. doi: 10.1016/j.tube.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Goodsell, D.S., Dickerson, R.E. Bending and curvature calculations in B-DNA. Nucleic Acids Res. 1994;22:5497–5503. doi: 10.1093/nar/22.24.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, G.A. The ACT domain: A small molecule binding domain and its role as a common regulatory element. J. Biol. Chem. 2006;281:33825–33829. doi: 10.1074/jbc.R600024200. [DOI] [PubMed] [Google Scholar]
- Grant, G.A., Schuller, D.J., Banaszak, L.J. A model for the regulation of D-3-phosphoglycerate dehydrogenase, a Vmax-type allosteric enzyme. Protein Sci. 1996;5:34–41. doi: 10.1002/pro.5560050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendickson, W.A., Ogata, C.M. Phase determination from multiwavelength anomalous diffraction measurements. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76074-9. [DOI] [PubMed] [Google Scholar]
- Holm, L., Sander, C. DALI: A network tool for protein structure comparison. Trends Biochem. Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- Hughes, J.D., Estep, P.W., Tavazoie, S., Church, G.M. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae . J. Mol. Biol. 2000;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- Ioerger, T.R., Sacchettini, J.C. The TEXTAL system: Artificial intelligence techniques for automated protein model building. Methods Enzymol. 2003;374:244–270. doi: 10.1016/S0076-6879(03)74012-9. [DOI] [PubMed] [Google Scholar]
- Kantardjieff, K.A., Hochtl, P., Segelke, B.W., Tao, F.M., Rupp, B. Concanavalin A in a dimeric crystal form: Revisiting structural accuracy and molecular flexibility. Acta Crystallogr. D Biol. Crystallogr. 2002;58:735–743. doi: 10.1107/s0907444901019588. [DOI] [PubMed] [Google Scholar]
- Koike, H., Ishijima, S.A., Clowney, L., Suzuki, M. The archaeal feast/famine regulatory protein: Potential roles of its assembly forms for regulating transcription. Proc. Natl. Acad. Sci. 2004;101:2840–2845. doi: 10.1073/pnas.0400109101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolling, R., Lother, H. AsnC: An autogenously regulated activator of asparagine synthetase A transcription in Escherichia coli . J. Bacteriol. 1985;164:310–315. doi: 10.1128/jb.164.1.310-315.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, H.J., Bennik, M.H., Demple, B., Ellenberger, T. Crystal structure of the Escherichia coli rob transcription factor complexed to DNA. Nat. Struct. Biol. 2000;7:424–430. doi: 10.1038/75213. [DOI] [PubMed] [Google Scholar]
- Landgraf, J.R., Wu, J., Calvo, J.M. Effects of nutrition and growth rate on Lrp levels in Escherichia coli . J. Bacteriol. 1996;1996:6930–6936. doi: 10.1128/jb.178.23.6930-6936.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard, P.M., Smits, S.H., Sedelnikova, S.E., Brinkman, A.B., de Vos, W.M., van der Oost, J., Rice, D.W., Rafferty, J.B. Crystal structure of the Lrp-like transcriptional regulator from the archaeon Pyrococcus furiosus . EMBO J. 2001;20:990–997. doi: 10.1093/emboj/20.5.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebel, R.O., Shorr, E., Richardson, H.B. The influence of foodstuffs upon the respiratory metabolism and growth of human tubercle bacilli. J. Bacteriol. 1933a;26:139–166. doi: 10.1128/jb.26.2.139-166.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebel, R.O., Shorr, E., Richardson, H.B. The influence of adverse conditions upon the respiratory metabolism and growth of human tubercle bacilli. J. Bacteriol. 1933b;26:167–200. doi: 10.1128/jb.26.2.167-200.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmberg, L.H., Hu, W.S., Sherman, D.H. Precursor flux control through targeted chromosomal insertion of the lysine epsilon-aminotransferase (lat) gene in cephamycin C biosynthesis. J. Bacteriol. 1993;175:6916–6924. doi: 10.1128/jb.175.21.6916-6924.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew, E., Zhi, J., Freundlich, M. Lrp is a direct repressor of the dad operon in Escherichia coli . J. Bacteriol. 1996;178:7234–7240. doi: 10.1128/jb.178.24.7234-7240.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, B.W. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- McRee, D.E. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- Miller, J.H. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1972. Experiments in molecular genetics. [Google Scholar]
- Mondragon, A., Subbiah, S., Almo, S.C., Drottar, M., Harrison, S.C. Structure of the amino-terminal domain of phage 434 repressor at 2.0 Å resolution. J. Mol. Biol. 1989;205:189–200. doi: 10.1016/0022-2836(89)90375-6. [DOI] [PubMed] [Google Scholar]
- Moreno-Hagelsieb, G., Collado-Vides, X. A powerful non-homology method for the prediction of operons in prokaryotes. Bioinformatics. 2002;18(Suppl. 1):S329–S336. doi: 10.1093/bioinformatics/18.suppl_1.s329. [DOI] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Muttucumaru, D.G., Roberts, G., Hinds, J., Stabler, R.A., Parish, T. Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis (Edinb.) 2004;84:239–246. doi: 10.1016/j.tube.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Neuwald, A.F., Liu, J.S., Lawrence, C.E. Gibbs motif sampling: Detection of bacterial outer membrane protein repeats. Protein Sci. 1995;4:1618–1632. doi: 10.1002/pro.5560040820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nou, X., Braaten, B., Kaltenbach, L., Low, D.A. Differential binding of Lrp to two sets of pap DNA binding sites mediated by Pap I regulates Pap phase variation in Escherichia coli . EMBO J. 1995;14:5785–5797. doi: 10.1002/j.1460-2075.1995.tb00267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z., Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. (1997);276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Ouhammouch, M., Geiduschek, E.P. An expanding family of archaeal transcriptional activators. Proc. Natl. Acad. Sci. 2005;102:15423–15428. doi: 10.1073/pnas.0508043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters, E., Thia-Toong, T.L., Gigot, D., Maes, D., Charlier, D. Ss-LrpB, a novel Lrp-like regulator of Sulfolobus solfataricus P2, binds cooperatively to three conserved targets in its own control region. Mol. Microbiol. 2004;54:321–336. doi: 10.1111/j.1365-2958.2004.04274.x. [DOI] [PubMed] [Google Scholar]
- Platko, J.V., Calvo, J.M. Mutations affecting the ability of Escherichia coli Lrp to bind DNA, activate transcription, or respond to leucine. J. Bacteriol. 1993;175:1110–1117. doi: 10.1128/jb.175.4.1110-1117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman, S., Hazra, R., Dascher, C.C., Husson, R.N. Transcription regulation by the Mycobacterium tuberculosis alternative σ factor SigD and its role in virulence. J. Bacteriol. 2004;186:6605–6616. doi: 10.1128/JB.186.19.6605-6616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick, G.M., Gould, R.O. Structure solution by iterative peaklist optimization and tangent expansion in space group P1. Acta Crystallogr. 1995;B51:432–446. [Google Scholar]
- Shrivastava, T., Kumar, S., Ramachandran, R. Cloning, expression, purification and crystallization of a transcriptional regulatory protein (Rv3291c) from Mycobacterium tuberculosis H37Rv. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1874–1876. doi: 10.1107/S0907444904016397. [DOI] [PubMed] [Google Scholar]
- Suzuki, M. Structure and function of the feast/famine regulatory proteins, FFRPs. Proc. Jpn. Acad. 2003;79B:274–289. [Google Scholar]
- Tani, T.H., Khodursky, A., Blumenthal, R.M., Brown, P.O., Matthews, R.G. Adaptation to famine: A family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. 2002;99:13471–13476. doi: 10.1073/pnas.212510999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapias, A., López, G., Ayora, S. Bacillus subtilis LrpC is a sequence-independent DNA-binding and DNA-bending protein which bridges DNA. Nucleic Acids Res. 2000;28:552–559. doi: 10.1093/nar/28.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T.C., Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaw, P., Sedelnikova, S.E., Muranova, T., Wiese, S., Ayora, S., Alonso, J.C., Brinkman, A.B., Akerboom, J., van der Oost, J., Rafferty, J.B. Structural insight into gene transcriptional regulation and effector binding by the Lrp/AsnC family. Nucleic Acids Res. 2006;34:1439–1449. doi: 10.1093/nar/gkl009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonrhein, C., Blanc, E., Roversi, P., Bricogne, G. Automated structure solution with autoSHARP. Methods Mol. Biol. 2006;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- Voskuil, M.I., Visconti, K.C., Schoolnik, G.K. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinb.) 2004;84:218–227. doi: 10.1016/j.tube.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Wang, Q., Calvo, J.M. Lrp, a global regulatory protein of Escherichia coli, binds co-operatively to multiple sites and activates transcription of ilvIH. J. Mol. Biol. 1993;229:306–318. doi: 10.1006/jmbi.1993.1036. [DOI] [PubMed] [Google Scholar]
- Wang, Q., Albert, F.G., Fitzgerald, D.J., Calvo, J.M., Anderson, J.N. Sequence determinants of DNA bending in the ilvlH promoter and regulatory region of Escherichia coli . Nucleic Acids Res. 1994;22:5753–5760. doi: 10.1093/nar/22.25.5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne, L.G., Hayes, L.G. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne, L.G., Sohaskey, C.D. Nonreplicating persistence of Mycobacterium tuberculosis . Annu. Rev. Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- Zinser, E.R., Kolter, R. Mutations enhancing amino acid catabolism confer a growth advantage in stationary phase. J. Bacteriol. 1999;181:5800–5807. doi: 10.1128/jb.181.18.5800-5807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]