Abstract
Acetyl-CoA carboxylase (ACC) catalyzes the first committed step in the synthesis of long-chain fatty acids. The crystal structure of the Escherichia coli carboxyltransferase component of ACC revealed an α2β2 subunit composition with two active sites and, most importantly, a unique zinc domain in each αβ pair that is absent in the eukaryotic enzyme. We show here that carboxyltransferase binds DNA. Half-maximal saturation of different single-stranded or double-stranded DNA constructs is seen at 0.5–1.0 μM, and binding is cooperative and nonspecific. The substrates (malonyl-CoA and biocytin) inhibit DNA:carboxyltransferase complex formation. More significantly, single-stranded DNA, double-stranded DNA, and heparin inhibit the reaction catalyzed by carboxyltransferase, with single-stranded DNA and heparin acting as competitive inhibitors. However, double-inhibition experiments revealed that both DNA and heparin can bind the enzyme in the presence of a bisubstrate analog (BiSA), and the binding of BiSA has a very weak synergistic effect on the binding of the second inhibitor (DNA or heparin) and vice versa. In contrast, DNA and heparin can also bind to the enzyme simultaneously, but the binding of either molecule has a strong synergistic effect on binding of the other. An important mechanistic implication of these observations is that the dual active sites of ACC are functionally connected.
Keywords: acetyl-CoA carboxylase, carboxyltransferase, biotin, zinc finger, DNA binding
Acetyl-CoA carboxylase is a multifunctional biotin-dependent enzyme that catalyzes the first committed and regulated step in fatty acid biosynthesis in bacteria via a two-step reaction (Scheme 1).
Scheme 1.
Biotin carboxylase catalyzes the first half-reaction, which is an ATP-dependent carboxylation of biotin to form carboxybiotin. The second half-reaction is catalyzed by carboxyltransferase, which transfers the carboxyl group from carboxybiotin to acetyl-CoA to generate malonyl-CoA. Biotin is covalently attached to the biotin carboxyl carrier protein (BCCP), and this complex is designated “Enzyme-biotin” in Scheme 1. In bacteria, the BCCP and the two enzymatic functions are encoded as three separate proteins (Cronan and Waldrop 2002). However, in eukaryotes, a single polypeptide chain comprises all three functional domains (Tanabe et al. 1975).
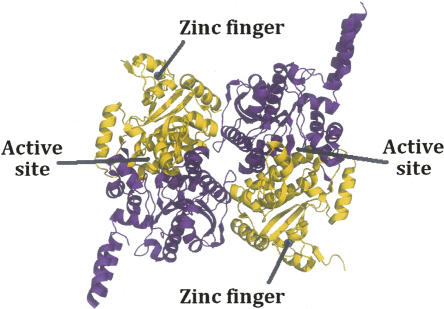
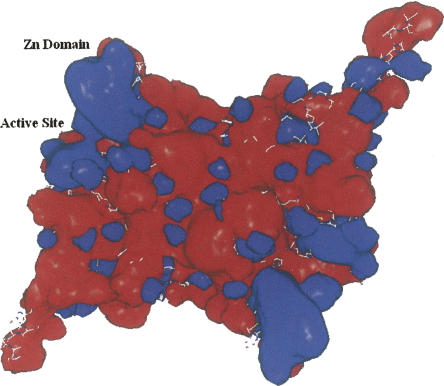
The recent crystal structures (Fig. 1) of carboxyltransferase from Escherichia coli and Staphylococcus aureus revealed a unique domain absent from eukaryotic homologs (Bilder et al. 2006). The structure confirmed the α2β2 subunit composition suggested by Lane and colleagues (Guchhait et al. 1974) and showed that the enzyme belongs to the crotonase superfamily (Gerlt and Babbitt 2001). The enzyme contains two active sites that lie at the interface of each of the αβ pairs (Fig. 1). The overall fold, not surprisingly, is similar to that of the carboxyltransferase domain from yeast (Zhang et al. 2003) and Streptomyces coelicolor (Diacovich et al. 2004). However, when the gene for the β-subunit of E. coli carboxyltransferase was cloned and sequenced 20 years ago, the authors noted the tandem C-X-X-C sequences separated by 15 residues located at the amino terminus and hypothesized that the protein may bind a metal ion (Bognar et al. 1987). The crystal structures of carboxyltransferase from S. aureus and E. coli, along with X-ray fluorescence studies, have, indeed, confirmed this prediction. The metal atom is zinc, and it forms part of a zinc motif that is unique to bacterial carboxyltransferase (Fig. 1). A space-filling representation reveals that the Zn domain forms part of a saddle-like structure (Fig. 2), and an electrostatic surface potential rendering shows that the inner face of the zinc finger domain has an electropositive surface potential, while most of the protein has an electronegative surface potential.
Figure 1.
Ribbon drawing of carboxyltransferase from S. aureus. (Purple) The α-chain; (gold) the β-chain. (Blue sphere in the β-chain) The zinc atom.
Figure 2.
Electrostatic surface potentials of the heterotetramer of carboxyltransferase. (Blue) A net positive charge; (red) a net negative charge. The surface potentials were generated with the program Grasp (Nicholls et al. 1991).
Given that Zn domains are commonly associated with DNA binding, and noting the favorable electrostatic potential, the aim of this study was to determine the ability of E. coli carboxyltransferase to bind DNA and characterize the effect of DNA binding on the enzymatic activity of carboxyltransferase. The results show that DNA, indeed, inhibits enzymatic activity; notably, the mode of binding reveals communication between the dual active sites of the functional protomers.
Results
DNA inhibits carboxyltransferase activity
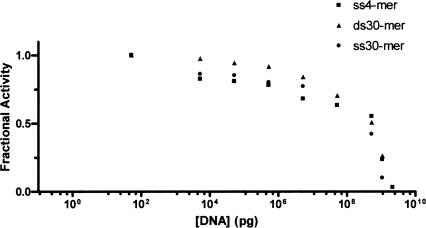
The zinc domain in bacterial carboxyltransferase belongs to the zinc ribbon class of zinc fingers (Krishna et al. 2003). Proteins that contain this type of zinc finger are frequently associated with DNA metabolism, such as the transcription factors TFIIS (Qian et al. 1993), TFIIB (Zhu et al. 1996), TFIIE (Okuda et al. 2004), several subunits from RNA polymerase II (Cramer et al. 2003), human ssDNA-binding protein RPA (Cochkareva et al. 2002), and bacteriophage T4 and T7 primases (Cha and Alberts 1986; Mendelman and Richardson 1991). Isolated zinc fingers like that in carboxyltransferase do not bind DNA tightly and recognize only three nucleotides (Wolfe et al. 2000). For example, T7 and T4 primases, recognize a preferred 3-nt sequence (Mendelman et al. 1999). Since carboxyltransferase contained an isolated zinc finger, it was assumed initially that DNA binding would be nonspecific. Therefore, to assess the ability of DNA to inhibit carboxyltransferase activity, random DNA sequences of varying lengths were examined. As shown in Figure 3, increasing concentrations of a 4-nt sequence composed of each of the four nucleotides, and a 30-nt PCR primer along with its complementary strand (i.e., the 30-bp DNA fragment) (Table 1) did, indeed, attenuate enzymatic activity comparably. It was not possible to test larger DNA fragments because the increased viscosity of the assay solution became prohibitive. However, viscosity or ionic strength is unlikely to account for the decrease in enzymatic activity due to the 4-nt and 30-nt DNA fragments since they inhibit to the same extent but would confer different viscosities and ionic strengths on the solutions. It is important to note that a thymidine dimer did not inhibit activity (data not shown), and that nucleotides have been previously reported not to affect activity (Polakis et al. 1973), suggesting that carboxyltransferase likely binds at least 3 nt. While the site of DNA binding has not been rigorously determined, we surmise that it includes the zinc finger given the overwhelming precedent for zinc fingers binding DNA.
Figure 3.
Dose-response curve for carboxyltransferase with both ssDNA and dsDNA. Initial velocity was measured at increasing amounts of DNA (4-nt ssDNA, 30-nt ssDNA, and 30-nt dsDNA) (Table 1). Malonyl-CoA was held constant at 0.1 mM, while biocytin was held constant at 5.0 mM.
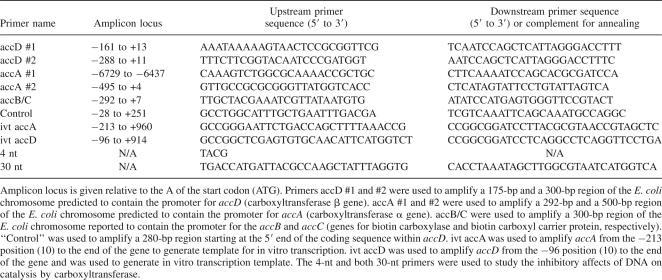
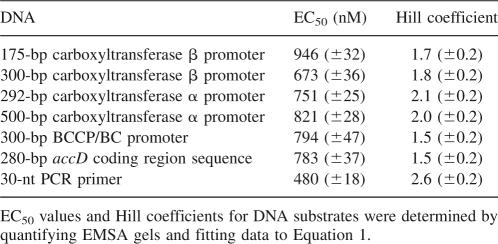
Table 1.
Primers used for amplification of substrate DNA or for enzyme inhibition assays
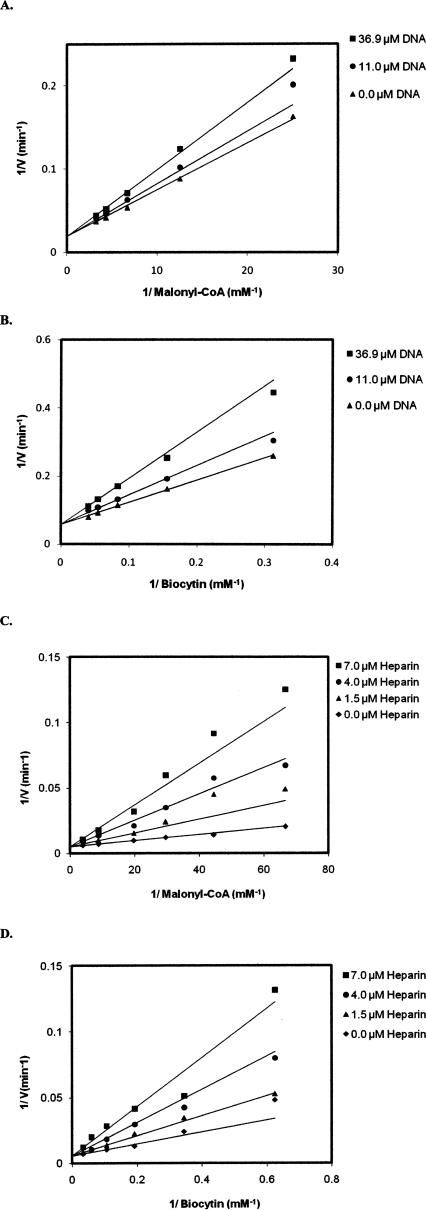
A single-stranded DNA substrate (30 nt upstream sequence) (Table 1) was used to examine the type of inhibition with respect to the substrates malonyl-CoA and biocytin.1 The 30-nt ssDNA exhibited competitive inhibition with respect to both malonyl-CoA and biocytin (Fig. 4A,B). Fitting the data to Equation 1 gave inhibition constants (K i) of 85 ± 10 μM with respect to malonyl-CoA and 34.2 ± 4.0 μM with respect to biocytin. The DNA mimic heparin was also found to inhibit enzymatic activity, and like DNA, exhibited competitive inhibition with respect to both substrates (Fig. 4C,D). The inhibition constants (K i) for heparin are 1.2 ± 0.1 μM with respect to malonyl-CoA and 2.2 ± 0.3 μM with respect to biocytin.
Figure 4.
Inhibition of carboxyltransferase by (A, B) DNA and (C, D) heparin. When malonyl-CoA was the variable substrate, biocytin was held constant at 5.0 mM, and when biocytin was the variable substrate, malonyl-CoA was held constant at 0.1 mM. The points are the reciprocal of the experimental velocities, and the lines are derived from the best fit of the data to Equation 1. The error on each velocity is <10%.
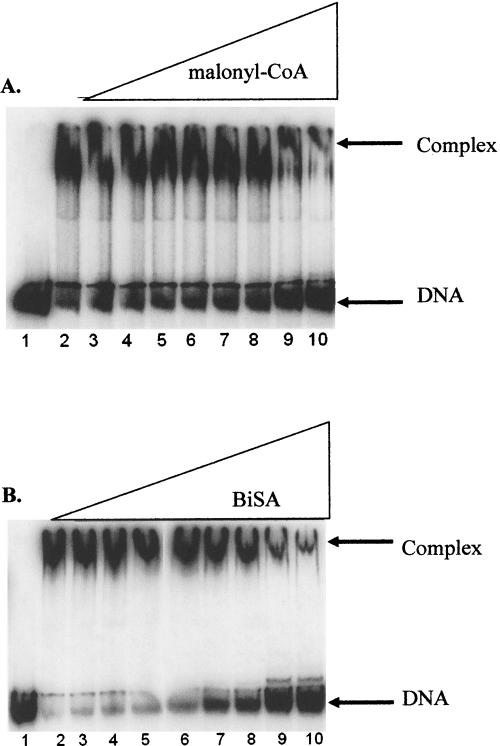
These inhibition data are consistent with electrophoretic mobility shift assays (EMSA), which show that increasing concentrations of malonyl-CoA inhibit formation of the carboxyltransferase–DNA complex (Fig. 5A). Moreover, a bisubstrate analog (BiSA) inhibitor of carboxyltransferase, in which coenzyme A is covalently attached to carboxybiotin (Levert and Waldrop 2002), was an even more efficient inhibitor of the enzyme–DNA complex (Fig. 5B). The substrate biocytin also inhibited formation of the carboxyltransferase–DNA complex, but only at very high concentrations (data not shown); because carboxyltransferase has an ordered addition of substrates with malonyl-CoA binding first (Blanchard and Waldrop 1998; Levert and Waldrop 2002), and given that biocytin exhibits substrate inhibition, at high concentrations biocytin binds in the malonyl-CoA-binding site and inhibits DNA binding.
Figure 5.
Inhibition of the formation of DNA:carboxyltransferase complex by malonyl-CoA and BiSA. All reactions contained 50 fmol of 300-bp carboxyltransferase β promoter DNA. (A) Reactions in lanes 2–10 have 800.0 nM carboxyltransferase; malonyl-CoA is titrated in reactions from lanes 3–10 (from 10.0 μM to 10.0 mM). (B) Reactions in lanes 2–10 have 800.0 nM carboxyltransferase; BISA is titrated in reactions in lanes 3–10 (from 2.5 μM to 2.5 mM).
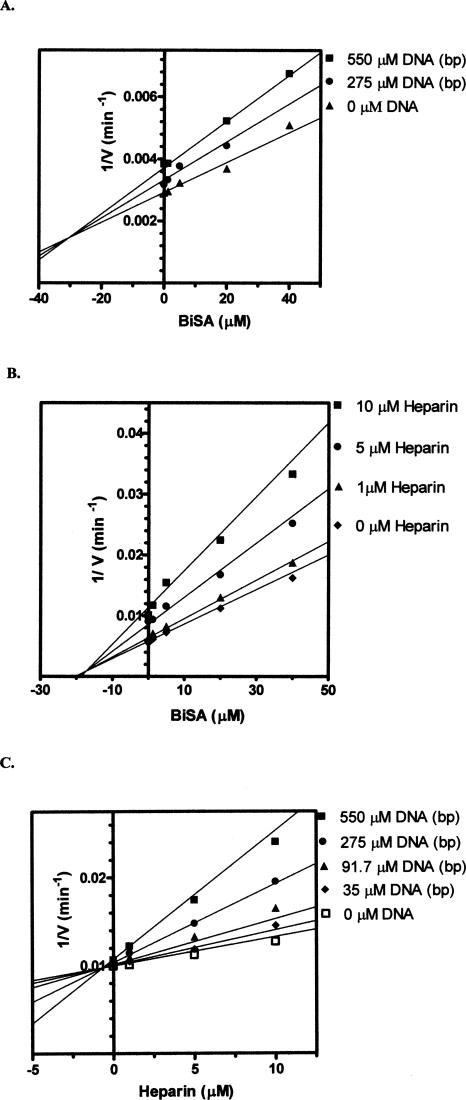
The competitive inhibition patterns indicate that DNA and heparin bind to the same enzyme form as the substrates malonyl-CoA and biocytin (namely, carboxyltransferase without substrates bound). In order to characterize the topological relationship between the DNA binding site and the active site (e.g., do the two sites overlap?), multiple inhibition studies were performed. Multiple inhibition experiments are carried out by measuring the initial velocity at increasing concentrations of one inhibitor while the second inhibitor is held constant. The substrate concentrations are held constant at subsaturating levels. The initial velocities are measured again at higher levels of the second inhibitor and then plotted as 1/velocity versus the concentration of the first inhibitor (sometimes referred to as a Yonetani-Theorell plot) (Yonetani and Theorell 1964). It is important to keep in mind that unlike the inhibition studies in Figure 4, A and B, where the substrate concentrations are extrapolated to infinity, the substrate concentrations in multiple inhibition experiments are held constant at subsaturating levels, which allows both inhibitors to bind to the enzyme. The first inhibitor for these studies was the bisubstrate analog (BiSA) used above for the EMSA assays. Since the substrate concentrations are held constant at subsaturating levels, BiSA could theoretically bind to an active site that already has DNA bound to it or to the other active site, which does not have DNA bound. Double inhibition of carboxyltransferase by either DNA or heparin at different fixed levels of BiSA (Fig. 6A,B) yielded intersecting patterns, which indicate that the two inhibitors can bind to the enzyme simultaneously. Thus, the binding site for DNA and the active site of the enzyme are topologically distinct, which suggests DNA binding at one αβ dimer, while BiSA binds to the other αβ dimer. This scenario is supported by fitting the data to Equation 2; β values of 0.50 and 0.85 were found for the DNA–BiSA inhibition and the heparin–BiSA inhibition, respectively. The β value is an indication of the interaction of the two inhibitors. Values of β >1 indicate that the binding of the two inhibitors interfere with one another, while a value of 1 indicates no interaction between the inhibitors. A β value <1 indicates synergism in the binding of the inhibitors. Thus, the binding of either DNA or heparin shows very weak synergism with the binding of BiSA and vice versa. Surprisingly, double-inhibition analysis with DNA and heparin also resulted in an intersecting pattern (Fig. 6C), indicating that DNA and heparin can bind to the enzyme simultaneously. However, the β value of 0.02 reflects a strong synergistic relationship in the binding of the two inhibitors. Given the presence of two Zn domains per protomer, the simplest explanation is that binding at one site enhances binding at the other.
Figure 6.
(A–C) Multiple inhibition patterns for DNA (ssDNA 30-nt upstream sequence) (Table 1), heparin, and BiSA. The points are the reciprocal of the experimental velocities, and the lines are derived from the best fit of the data to Equation 2. Malonyl-CoA was held constant at 0.1 mM, while biocytin was held constant at 5.0 mM.
DNA binding by carboxyltransferase
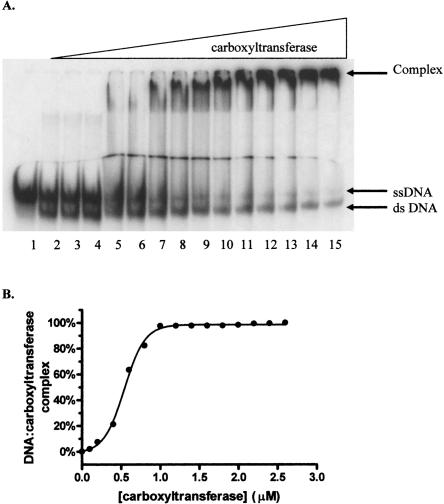
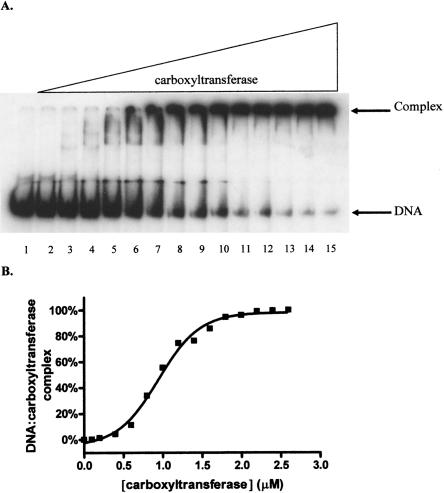
Since the multiple inhibition studies showed that DNA and the DNA mimic heparin exhibit synergistic binding to carboxyltransferase, DNA binding was characterized using EMSA. As shown in Figure 7A, carboxyltransferase binds 30-nt ssDNA in preference to duplex as evidenced by the disappearance of free ssDNA and the concomitant formation of a complex that fails to migrate from the well of the gel. When carboxyltransferase was incubated with only ssDNA and the region on the gel from the slowest migrating complex to the free nucleic acid was considered as the protein–DNA complex, quantification of the data shows that ssDNA binds to carboxyltransferase in a cooperative fashion (Fig. 7B), with a half-maximal saturation of 480 ± 18 nM (Table 2). That complexes formed at higher carboxyltransferase concentrations fail to migrate from the well suggests the formation of aggregates, consistent with the presence of two zinc domains, the presumed sites of DNA binding.
Figure 7.
Electrophoretic analysis of DNA titrated with carboxyltransferase. (A, lane 1) ssDNA (50.0 fmol). The remaining reactions contained a 50-fmol ssDNA with one-half molar ratio complementary strand, annealed by cooling over 3 h from 95°C to 0°C. Reactions in lanes 3–15 contain 0.1, 0.2, 0.4, 0.6, 0.8, 1.0, 1.2, 1.4, 1.6, 1.8, 2.0, 2.2, and 2.4 μM carboxyltransferase. (B) Binding isotherm for carboxyltransferase binding to ssDNA in the absence of dsDNA (30-nt upstream primer sequence) (Table 1). The line represents the best fit of the data to Equation 3.
Table 2.
Half maximal values for carboxyltransferase binding to substrate DNA molecules
Since the four components (i.e., biotin carboxylase, BCCP, and the α and β subunits of carboxyltransferase) of the E. coli acetyl-CoA carboxylase are likely produced in a defined, stoichiometric ratio, and since expression of the genes has been reported to be directly correlated to cellular growth rates (Li and Cronan 1993), we explored the possibility that carboxyltransferase may contribute to regulation of any of the genes encoding the acetyl-CoA carboxylase subunits.
Carboxyltransferase binds a 175-bp DNA containing the accD promoter (i.e., the gene encoding the β-subunit) (Li and Cronan 1993), as evidenced by the formation of a complex that fails to migrate from the well of the gel (Fig. 8A). As with the 30-nt ssDNA, binding was cooperative with a half-maximal saturation of 946 ± 32 nM (Table 2). This confirms that cooperative binding was not unique to ssDNA. In fact, carboxyltransferase bound cooperatively to DNA corresponding to the promoter regions of the gene coding for the α-subunit (accA), and the operon that includes the genes for biotin carboxylase and the biotin carboxyl carrier protein as well as a 280-bp DNA representing the accD-coding region. The comparable half-maximal saturation for all the DNA sequences examined (Table 2) suggests that carboxyltransferase binds DNA with low affinity and nonspecifically.
Figure 8.
Electrophoretic analysis of 175-bp dsDNA from promoter region of accD (carboxyltransferase β-subunit) titrated with carboxyltransferase. (A) Reactions contain 50.0 fmol of DNA. Reactions in lanes 2–15 contain 0.1, 0.2, 0.4, 0.6, 0.8, 1.0, 1.2, 1.4, 1.6, 1.8, 2.0, 2.2, 2.4, and 2.6 μM carboxyltransferase. (B) Binding isotherm for carboxyltransferase binding to 175-bp dsDNA. The line represents the best fit of the data to Equation 3.
While we cannot rule out that carboxyltransferase may be recruited to specific DNA sites by association with other factors, it is important to note that addition of biotin carboxylase or the biotin carboxyl carrier protein (biotinylated and unbiotinylated) did not increase the affinity of carboxyltransferase for duplex or single-stranded DNA, suggesting that the other components of acetyl-CoA carboxylase do not confer high-affinity DNA binding (data not shown). Moreover, while acyl-ACP has been reported to be a feedback inhibitor of carboxyltransferase (Davis and Cronan 2001), we found that increasing amounts up to 100 μM had no effect on formation of the DNA–carboxyltransferase complex (data not shown). In addition, MfpA, a protein from Mycobacterium tuberculosis that provides resistance to fluoroquinolones by mimicking DNA and binding to DNA gyrase (Hegde et al. 2005), also did not inhibit formation of the carboxyltransferase–DNA complex (data not shown). Lastly, considering the potential for carboxyltransferase looping DNA and the presence of AT-rich sequences weakly resembling consensus binding sites for integration host factor (IHF) (Freundlich et al. 1992; Hales et al. 1994, 1996), we assessed the ability of IHF to enhance carboxyltransferase–DNA complex formation. IHF did not increase the affinity of carboxyltransferase for either of the promoter regions of the genes for the α- and β-subunits (data not shown). Taken together, the results indicate that carboxyltransferase exhibits nonspecific, cooperative DNA binding.
Discussion
DNA-binding enzymes
Few enzymes that catalyze a reaction in intermediary metabolism also bind DNA or RNA. Examples include PutA (proline utilization A) (Brown and Wood 1992); BirA or biotin ligase, which attaches biotin to the biotin carboxyl carrier protein (Beckett 2005); the plant cysteine protease LeCp (Matarasso et al. 2005); Arg 5,6, which is involved in arginine biosynthesis (Hall et al. 2004); Ilv5p, which catalyzes a reaction in branched-chain amino acid biosynthesis (Bateman et al. 2002a); and iron regulatory protein 1 (IRP1), which binds mRNA and functions as aconitase (Walden et al. 2006). These proteins all have dual functions; they either act as enzymes or as nucleic acid binding proteins, and, most importantly, these two functions can be separated. For example, the DNA-binding domain of PutA can be expressed and purified without the enzymatic domain, and it still binds DNA (Gu et al. 2004). For Ilv5p, either the DNA-binding domain or the enzymatic domain can be inactivated without affecting the activity of the other domain (Bateman et al. 2002b). What distinguishes carboxyltransferase from these enzymes is that catalysis and DNA binding are inextricably linked and that DNA binding is cooperative.
The mode of DNA binding suggests communication between the dual active sites
DNA and heparin are competitive inhibitors with respect to both substrates malonyl-CoA and biocytin. The competitive inhibition patterns indicate that saturating levels of either substrate prevent DNA binding. This makes intuitive sense for malonyl-CoA; the β-subunit contains the zinc domain, the presumed DNA binding site, in close proximity to the malonyl-CoA-binding site. Biocytin, however, binds to the α-subunit. Given that DNA is a polymer, the competitive inhibition pattern with respect to biocytin is likely due to a steric effect, where the polymeric DNA blocks access to the biocytin-binding site. Inhibition of carboxyltransferase by DNA is consistent with EMSA data showing that malonyl-CoA as well as a bisubstrate analog prevent formation of the DNA–protein complex. Thus, enzymatic activity and DNA binding are not separable functions.
The cooperative binding of DNA to carboxyltransferase is likely a manifestation of two separate physical phenomena. Carboxyltransferase has two zinc domains diametrically opposed, and DNA has multiple binding sites. Accretion of carboxyltransferase mediated by protein–protein interactions could account for the observed cooperativity. Second, the binding of DNA to one site on carboxyltransferase could increase the affinity for DNA binding to the second site. Significantly, evidence for intersubunit communication is also seen in the multiple inhibition experiments with DNA and heparin (Fig. 6) where the β value, which reports on the degree of synergism in the binding of the two inhibitors, is well below 1 (0.02). This synergistic binding of DNA to carboxyltransferase could certainly contribute to the sigmoidal curve observed for DNA binding. Taken together, our data suggest functional communication between the dual active sites of carboxyltransferase. Synergistic substrate binding to carboxyltransferase in vivo would promote rapid initiation of fatty acid biosynthesis under favorable conditions.
Does carboxyltransferase bind DNA in vivo?
The physical parameters derived from this work can be used to predict whether carboxyltransferase might bind DNA in vivo. A 3-nt binding site is assumed, which is consistent with other zinc finger-binding sites (Wolfe et al. 2000). However, based on the dimensions of carboxyltransferase, a single molecule of enzyme would occlude ∼30 bp, which would result in 1.5 × 105 or 0.75 mM potential binding sites for carboxyltransferase. The intracellular concentration of carboxyltransferase is ∼500 nM (Guchhait et al. 1974). Using 0.8 μM as the binding constant of DNA to carboxyltransferase, the fraction of carboxyltransferase bound to DNA intracellularly can be calculated with the following equation (Segel 1975):
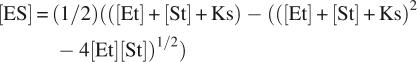 |
where Et is the concentration of carboxyltransferase, St is the concentration of DNA-binding sites, and Ks is the dissociation constant for DNA binding to carboxyltransferase. Assuming that the entire E. coli genome is accessible to carboxyltransferase, the calculation suggests that all of the enzyme would be bound to DNA in vivo. However, most of the genomic DNA in E. coli is compacted by proteins (Minsky 2004) and would be inaccessible to carboxyltransferase, yet recalculation assuming only 10% of the genomic DNA available for binding to carboxyltransferase also suggests that all of the carboxyltransferase in E. coli would be bound to DNA. Even if the K d were 10-fold higher in vivo (given the higher intracellular salt concentrations), 90% of the enzyme would still be bound to DNA, again assuming that only 10% of the genomic DNA is available for binding.
What might be the consequences of a significant fraction of carboxyltransferase being bound to DNA? The observation that both substrates inhibit DNA binding is very important when considering how carboxyltransferase could function as a critical enzyme for membrane biogenesis. It is tempting to speculate that during the stationary phase of E. coli growth, where nutrients are limited, carboxyltransferase functions as a nucleoid-associated protein (Drlica and Rouvieìre-Yaniv 1987) to compact and protect the chromosome from damage. During the growth phase when nutrients are abundant, the levels of substrates, most notably acetyl-CoA, increase dramatically and compete with DNA for binding to carboxyltansferase, and the enzyme functions to synthesize fatty acids for cell membrane assembly. During stationary phase, when carboxyltransferase activity is low, association with nucleic acids (ssDNA and dsDNA) may also prevent futile activity and/or the sequestration of cellular metabolites.
Materials and Methods
Restriction enzymes, dNTPs, and T4 DNA ligase were purchased from New England Biolabs. Pfu Turbo DNA polymerase was from Stratagene. Primers for PCR and oligonucleotides used to generate dsDNA and ssDNA used as inhibitors were obtained from MWG BioTech. Heparin, coupling enzymes, and NAD were from Sigma/Aldrich. The bisubstrate analog (BiSA) inhibitor of carboxyltransferase was synthesized according to the method of Levert and Waldrop (2002). [γ-32P]ATP was from PerkinElmer. QIAquick Gel Extraction Kit and DNeasy Tissue Kit were from QIAGEN, Inc. IHF was obtained as described by Grove et al. (1996). MfpA was kindly provided by John Blanchard, Albert Einstein College of Medicine, New York.
Purification and enzymatic assay of carboxyltransferase
Carboxyltransferase was purified from E. coli transformed with an overexpression plasmid containing a mini-operon with genes for the α- and β-subunits of the enzyme (Blanchard and Waldrop 1998).
Carboxyltransferase activity was measured in the reverse direction with a spectrophotometric assay in which the production of acetyl-CoA was coupled to the combined citrate synthase-malate dehydrogenase reaction requiring NAD+ reduction (Blanchard and Waldrop 1998). NADH formation was followed spectrophotometrically at 340 nm using a Uvikon 810 (Kontron Instruments) spectrophotometer interfaced to a PC equipped with a data acquisition program. Since the crystal structure showed that carboxyltransferase contains two active sites, the initial velocities were calculated per active site using a molecular weight of 68.5 kDa for each αβ dimer (i.e., active site). Control experiments showed that neither DNA nor heparin at the highest concentration used in Figure 4 inhibited the coupling enzymes citrate synthase or malate dehydrogenase.
DNA substrates
The promoter regions for the α (292-bp and 500-bp fragments) and β (175-bp and 300-bp fragments) genes of carboxyltransferase, the promoter region for the biotin carboxylase/biotin carboxyl carrier protein (300-bp fragment), and a 280-bp fragment encompassing part of the accD coding region were PCR-amplified using the primers listed in Table 1. E. coli (JM109; New England Biolabs) chromosomal DNA isolated via DNeasy Tissue kit was used as the template. The PCR products were electrophoresed, and their concentration was assessed by densitometry by comparing sample intensity to that of similar-sized molecular weight standards. The PCR products were 5′-end-labeled with T4 polynucleotide kinase and [γ-32P]ATP and purified via QIAquick Gel Extraction Kit, assuming 90% sample recovery.
Electrophoretic mobility shift assay (EMSA)
The reactions (total reaction volume of 10 μL) included 50 fmol of DNA, with carboxyltransferase titrated from 0 to 2.6 μM. The binding buffer was 20 mM Tris-HCl (pH 8), 0.1 mM Na2EDTA, 0.075% BRIJ58, 50 mM NaCl, 5 mM MgCl2, 50 μg/mL BSA, and 4% (v/v) glycerol. Reactions were incubated on ice for 0.5 h. Samples were resolved on prerun 6.0% (w/v) native polyacrylamide gels (39:1 acrylamide:bisacrylamide) at room temperature in TBE buffer (45 mM Tris borate at pH 8.3, 1.25 mM Na2EDTA), and samples were loaded with the power on. After 2.0 h of electrophoresis, the gels were dried and complexes were visualized by PhosphorImaging and quantified using ImageQuant TL (GE Healthcare). For calculating percent complex formation, the region on the gel from the slowest migrating complex to the free nucleic acid was considered as complex. The percent complex was calculated as complex intensity divided by the total intensity (or total nucleic acid) in a given lane. Binding assays were carried out in duplicate. Binding isotherms are from representative EMSAs, with <17.5% variation between experiments.
Inhibitors of DNA binding by carboxyltransferase were analyzed by measuring percent complex versus inhibitor concentration. A competition assay was carried out similar to the binding assay for affinity determination (above), except the carboxyltransferase was held constant and the inhibitor concentration varied. The order of addition for each of the regents in the competition assay is as follows: buffer, carboxyltransferase, inhibitor molecule(s), and labeled DNA.
Data analysis
Competitive inhibition data were fitted to Equation 1, using the programs of Cleland (1979). In Equation 1, v is the initial velocity, V m is the maximal velocity, A is the substrate concentration, I is the concentration of inhibitor, K m is the Michaelis constant, K i is the inhibition constant. Data for multiple inhibitions were fitted to Equation 2, where v is the initial velocity, I and J are the concentrations of the two inhibitors, v 0 is the velocity in the absence of inhibitors, Ki and Kj are the apparent dissociation constants for the two inhibitors, and β is a measure of the degree of interaction of the two inhibitors (Cleland 1990). The binding isotherms for carboxyltransferase binding to DNA were analyzed by fitting the data to Equation 3 to calculate the concentration of protein resulting in half-maximal saturation (EC50). In Equation 3, Y equals the fractional complex, while min is the minimum value of Y and max is the maximum value of Y. x is the concentration of carboxyltransferase, and n is the Hill coefficient.
Acknowledgments
We thank John Blanchard of Albert Einstein College of Medicine for providing MfpA. We also thank W.W. Cleland of the University of Wisconsin for help in fitting the multiple inhibition data. Special thanks to Nabil Thalji for making Figure 2. B.K.B. was supported by the National Science Foundation under Grant No. DGE-9987603.
Footnotes
The catalytic activity of carboxyltransferase from E. coli is often measured in the reverse, or nonphysiological, direction with a facile spectrophotometric continuous assay. The assay links the production of acetyl-CoA to the reduction of NAD+ by the coupled reactions of citrate synthase and malate dehydrogenase (Blanchard and Waldrop 1998). Biocytin is preferred over biotin because biocytin produces a maximal velocity three orders of magnitude greater than does biotin (Blanchard and Waldrop 1998). Biocytin is a biotin molecule with a lysine appended to the carboxyl group of the valeric acid side chain via an amide linkage at the ε-amino group.
Reprint requests to: Grover L. Waldrop, Department of Biological Sciences, Room 206 Life Sciences Building, Louisiana State University, Baton Rouge, LA 70803, USA; e-mail: gwaldro@lsu.edu; fax: (225) 578-7258.
Abbreviations: ACC, acetyl-CoA carboxylase; BCCP, biotin carboxyl carrier protein; IHF, integration host factor; BiSA, bisubstrate analog; EMSA, electrophoretic mobility shift assay.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073186408.
References
- Bateman, J.M., Iacovino, M., Perlman, P.S., Butow, R.A. Mitochondrial DNA instability mutants of the bifunctional protein Ilv5p have altered organization in mitochondria and are targeted for degradation by Hsp78 and Pim1p protease. J. Biol. Chem. 2002a;277:47946–47953. doi: 10.1074/jbc.M209071200. [DOI] [PubMed] [Google Scholar]
- Bateman, J.M., Perlman, P.S., Butow, R.A. Mutational bisection of the mitochondrial DNA stability and amino acid biosynthetic functions of Ilv5p of budding yeast. Genetics. 2002b;161:1043–1052. doi: 10.1093/genetics/161.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett, D. The Escherichia coli biotin regulatory system: A transcriptional switch. J. Nutr. Biochem. 2005;16:411–415. doi: 10.1016/j.jnutbio.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Bilder, P., Lightle, S., Bainbridge, G., Ohren, J., Finzel, B., Sun, F., Holley, S., Al-Kassim, L., Spessard, C. The structure of the carboxyltransferase component of acetyl-coA carboxylase reveals a zinc-binding motif unique to the bacterial enzyme. Biochemistry. 2006;45:1712–1722. doi: 10.1021/bi0520479. [DOI] [PubMed] [Google Scholar]
- Blanchard, C.Z., Waldrop, G.L. Overexpression and kinetic characterization of the carboxyltransferase component of acetyl-CoA carboxylase. J. Biol. Chem. 1998;273:19140–19145. doi: 10.1074/jbc.273.30.19140. [DOI] [PubMed] [Google Scholar]
- Bognar, A.L., Osborne, C., Shane, B. Primary structure of the Escherichia coli folC gene and its folylpolyglutamate synthetase–dihydrofolate synthetase product and regulation of expression by an upstream gene. J. Biol. Chem. 1987;262:12337–12343. [PubMed] [Google Scholar]
- Brown, E., Wood, J.M. Redesigned purification yields a fully functional PutA protein dimer from Escherichia coli . J. Biol. Chem. 1992;267:13086–13092. [PubMed] [Google Scholar]
- Cha, T.A., Alberts, B.M. Studies of the DNA helicase–RNA primase unit from bacteriophage T4. A trinucleotide sequence on the DNA template starts RNA primer synthesis. J. Biol. Chem. 1986;261:7001–7010. [PubMed] [Google Scholar]
- Cleland, W.W. Statistical analysis of enzyme kinetic data. Methods Enzymol. 1979;63:103–138. doi: 10.1016/0076-6879(79)63008-2. [DOI] [PubMed] [Google Scholar]
- Cleland, W.W. Steady-state kinetics. In: Sigman D.S., Boyer P.D., editors. The enzymes. Academic Press; San Diego, CA: 1990. pp. 117–119. [Google Scholar]
- Cochkareva, E., Korolev, S., Lees-Miller, S.P., Bochkarev, A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 2002;21:1855–1863. doi: 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer, P., Bushnell, D.A., Kornberg, R.D. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2003;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- Cronan J.E., Jr, Waldrop, G.L. Multi-subunit acetyl-CoA carboxylases. Prog. Lipid Res. 2002;41:407–435. doi: 10.1016/s0163-7827(02)00007-3. [DOI] [PubMed] [Google Scholar]
- Davis, M.S., Cronan J.E., Jr Inhibition of Escherichia coli acetyl coenzyme A carboxylase by acyl-acyl carrier protein. J. Bacteriol. 2001;183:1499–1503. doi: 10.1128/JB.183.4.1499-1503.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diacovich, L., Mitchell, D.L., Pham, H., Gago, G., Melgar, M.M., Khosla, C., Gramajo, H., Tsai, S.C. Crystal structure of the β-subunit of acyl-CoA carboxylase: Structure-based engineering of substrate specificity. Biochemistry. 2004;43:14027–14036. doi: 10.1021/bi049065v. [DOI] [PubMed] [Google Scholar]
- Drlica, K., Rouvieìre-Yaniv, J. Histone-like proteins of bacteria. Microbiol. Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freundlich, M., Ramani, N., Mathew, E., Sirko, A., Tsui, P. The role of integration host factor in gene expression in Escherichia coli . Mol. Microbiol. 1992;6:2557–2563. doi: 10.1111/j.1365-2958.1992.tb01432.x. [DOI] [PubMed] [Google Scholar]
- Gerlt, J.A., Babbitt, P.C. Divergent evolution of enzymatic function: Mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu. Rev. Biochem. 2001;70:209–246. doi: 10.1146/annurev.biochem.70.1.209. [DOI] [PubMed] [Google Scholar]
- Grove, A., Galeone, A., Mayol, L., Geiduschek, E.P. Localized DNA flexibility contributes to target site selection by DNA-bending proteins. J. Mol. Biol. 1996;260:120–125. doi: 10.1006/jmbi.1996.0386. [DOI] [PubMed] [Google Scholar]
- Gu, D., Zhou, Y., Kallhoff, V., Baban, B., Tanner, J.J., Becker, D.F. Identification and characterization of the DNA-binding domain of the multifunctional PutA flavoenzyme. J. Biol. Chem. 2004;279:31171–31176. doi: 10.1074/jbc.M403701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guchhait, R.B., Polakis, S.E., Dimroth, P., Stoll, E., Moss, J., Lane, M.D. Acetyl coenzyme A carboxylase system of Escherichia coli. Purification and properties of the biotin carboxylase, carboxyltransferase, and carboxyl carrier protein components. J. Biol. Chem. 1974;249:6633–6645. [PubMed] [Google Scholar]
- Hales, L.M., Gumport, R.I., Gardner, J.F. Determining the DNA sequence elements required for binding integration host factor to two different target sites. J. Bacteriol. 1994;176:2999–3006. doi: 10.1128/jb.176.10.2999-3006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales, L.M., Gumport, R.I., Gardner, J.F. Examining the contribution of a dA+dT element to the conformation of Escherichia coli integration host factor–DNA complexes. Nucleic Acids Res. 1996;24:1780–1786. doi: 10.1093/nar/24.9.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, D.A., Zhu, H., Zhu, X., Royce, T., Gerstein, M., Snyder, M. Regulation of gene expression by a metabolic enzyme. Science. 2004;306:482–484. doi: 10.1126/science.1096773. [DOI] [PubMed] [Google Scholar]
- Hegde, S.S., Vetting, M.W., Roderick, S.L., Mitchenall, L.A., Maxwell, A., Takiff, H.E., Blanchard, J.S. A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science. 2005;308:1480–1483. doi: 10.1126/science.1110699. [DOI] [PubMed] [Google Scholar]
- Krishna, S.S., Majumdar, I., Grishin, N.V. Structural classification of zinc fingers. Nucleic Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levert, K.L., Waldrop, G.L. A bisubstrate analog inhibitor of the carboxyltransferase component of acetyl-CoA carboxylase. Biochem. Biophys. Res. Commun. 2002;291:1213–1217. doi: 10.1006/bbrc.2002.6576. [DOI] [PubMed] [Google Scholar]
- Li, S.J., Cronan J.E., Jr Growth rate regulation of Escherichia coli acetyl coenzyme A carboxylase, which catalyzes the first committed step of lipid biosynthesis. J. Bacteriol. 1993;175:332–340. doi: 10.1128/jb.175.2.332-340.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarasso, N., Schuster, S., Avni, A. A novel plant cysteine protease has a dual function as a regulator of 1-aminocyclopropane-1-carboxylic acid synthase gene expression. Plant Cell. 2005;17:1205–1216. doi: 10.1105/tpc.105.030775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelman, L.V., Richardson, C.C. Requirements for primer synthesis by bacteriophage T7 63-kDa gene 4 protein. Roles of template sequence and T7 56-kDa gene 4 protein. J. Biol. Chem. 1991;266:23240–23250. [PubMed] [Google Scholar]
- Mendelman, L., Kusakabe, T., Hine, A., Hyberts, S.G., Richardson, C.C. The Cys4 zinc finger of bacteriophage T7 primase in sequence-specific single-stranded DNA recognition. Proc. Natl. Acad. Sci. 1999;96:4295–4300. doi: 10.1073/pnas.96.8.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minsky, A. Information content and complexity in the high-order organization of DNA. Annu. Rev. Biophys. Biomol. Struct. 2004;33:317–342. doi: 10.1146/annurev.biophys.33.110502.133328. [DOI] [PubMed] [Google Scholar]
- Nicholls, A., Sharp, K., Honig, B. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Okuda, M., Tanaka, A., Arai, Y., Satoh, M., Okamura, H., Nagadoi, A., Hanaoka, F., Okduma, Y., Nishimura, Y. A novel zinc finger structure in the large subunit of human general transcription factor TFIIE. J. Biol. Chem. 2004;279:51395–51403. doi: 10.1074/jbc.M404722200. [DOI] [PubMed] [Google Scholar]
- Polakis, S.E., Guchhait, R.B., Lane, M.D. Stringent control of fatty acid synthesis in Escherichia coli . J. Biol. Chem. 1973;248:7957–7966. [PubMed] [Google Scholar]
- Qian, X., Gozani, S., Yoon, H., Jeon, C., Agarwal, K., Weiss, M.A. Novel zinc finger motif in the basal transcriptional machinery: Three-dimensional NMR studies of the nucleic acid binding domain of transcriptional elongation factor TFIIS. Biochemistry. 1993;32:9944–9959. doi: 10.1021/bi00089a010. [DOI] [PubMed] [Google Scholar]
- Segel, I.H. Enzyme kinetics: Behavior and analysis of rapid equilibrium and steady-state enzyme systems. Wiley; New York: 1975. p. 73. [Google Scholar]
- Tanabe, T., Wada, K., Okazaki, T., Numa, S. Acetyl-coenzyme-A carboxylase from rat liver. Subunit structure and proteolytic modification. Eur. J. Biochem. 1975;57:15–24. doi: 10.1111/j.1432-1033.1975.tb02272.x. [DOI] [PubMed] [Google Scholar]
- Walden, W.E., Selezneva, A.I., Dupuy, J., Volbeda, A., Fontecilla-Camps, J.C., Theil, E.C., Volz, K. Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA. Science. 2006;314:1903–1908. doi: 10.1126/science.1133116. [DOI] [PubMed] [Google Scholar]
- Wolfe, S.A., Nekludova, L., Pabo, C.O. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- Yonetani, T., Theorell, H. Studies on liver alcohol hydrogenase complex 3. Multiple inhibition kinetics in the presence of two competitive inhibitors. Arch. Biochem. Biophys. 1964;106:243–251. doi: 10.1016/0003-9861(64)90184-5. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Yang, Z., Shen, Y., Tong, L. Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase. Science. 2003;299:2064–2067. doi: 10.1126/science.1081366. [DOI] [PubMed] [Google Scholar]
- Zhu, W., Zeng, Q., Colangelo, C.M., Lewis, L.M., Summers, M.F., Scott, R.A. The N-terminal domain of TFIIB from Pyrococcus furiosus forms a zinc ribbon. Nat. Struct. Biol. 1996;3:122–124. doi: 10.1038/nsb0296-122. [DOI] [PubMed] [Google Scholar]