Abstract
Secreted and transmembrane proteins play an essential role in intercellular communication during the development of multicellular organisms. Because only a small number of these genes have been characterized, we developed a screen for genes encoding extracellular proteins that are differentially expressed during Drosophila embryogenesis. Our approach utilizes a new method for screening large numbers of cDNAs by whole-embryo in situ hybridization. The cDNA library for the screen was prepared from rough endoplasmic reticulum-bound mRNA and is therefore enriched in clones encoding membrane and secreted proteins. To increase the prevalence of rare cDNAs in the library, the library was normalized using a method based on cDNA hybridization to genomic DNA-coated beads. In total, 2,518 individual cDNAs from the normalized library were screened by in situ hybridization, and 917 of these cDNAs represent genes differentially expressed during embryonic development. Sequence analysis of 1,001 cDNAs indicated that 811 represent genes not previously described in Drosophila. Expression pattern photographs and partial DNA sequences have been assembled in a database publicly available at the Berkeley Drosophila Genome Project website (http://fruitfly.berkeley.edu). The identification of a large number of genes encoding proteins involved in cell–cell contact and signaling will advance our knowledge of the mechanisms by which multicellular organisms and their specialized organs develop.
A major goal of developmental biology is to elucidate the molecular mechanisms that govern cell–cell interactions in higher eukaryotes. Genetic analysis of development in Drosophila has proven to be a powerful approach for studying these mechanisms. For example, most of the genes known to be involved in the hedgehog (1, 2), dpp/BMP (3), and Wnt (4) signaling pathways were identified through classical genetic screens in Drosophila. The characterization of these genes and their vertebrate homologs has greatly advanced our understanding of the cell signaling pathways that regulate development.
Genetic screens, however, have significant limitations. Genes with subtle loss-of-function phenotypes or genes whose function can be compensated for by other genes or pathways are unlikely to be found. These two classes of genes may represent the majority of genes in Drosophila, since it is estimated that two-thirds of Drosophila genes are not required for viability (5). In addition, screens designed to identify specific phenotypic defects often do not recover genes with pleiotropic roles during development, since the requirement for gene function in one developmental process can mask its requirement in another.
To identify all classes of developmentally important genes, expression-based and other molecular screens are needed to supplement classical genetic screens. In Drosophila, the most productive of these screens to date have utilized P element-based enhancer traps (6–9), but P element insertion is not random and enhancer trap screens are biased toward identifying genes that are favored for insertion by P elements (10). Other expression-based screens to specifically identify extracellular proteins have involved generating monoclonal antibodies against crude membrane preparations and screening by immunostaining of embryos (11, 12). Unfortunately, antibody screens are biased toward identifying the most abundant or highly immunogenic proteins and thus typically identify only a small subset of proteins.
We present a large-scale screen for genes encoding secreted and transmembrane proteins that are expressed in specific tissue or cell types during embryonic development in Drosophila. The approach combines a cDNA library enriched for genes encoding extracellular proteins with a high throughput whole-embryo in situ hybridization procedure and subsequent sequence analysis. The results have been compiled in a publicly available database.
MATERIALS AND METHODS
All protocols used in this study are available in a more detailed form at http://fruitfly.berkeley.edu.
RNA Isolation from Rough Endoplasmic Reticulum.
Rough endoplasmic reticulum membranes or rough microsomes (RMs) were isolated from 10 g of 8- to 16-hr (25°C) embryos using a sucrose gradient sedimentation procedure (13, 14) with some modifications. Poly(A)+ RNA was purified from the RM RNA preparation using the Poly(A) Select kit (Promega).
cDNA Library Construction.
A directionally cloned RM cDNA library was prepared from RM poly(A)+ RNA using standard techniques (15), except that the RNA was annealed with a Pst-T15 primer/adaptor (5′-CACCTTGTCTCACTGCAGT15) and the first-strand cDNA was synthesized in the presence of 5-methyl dCTP (Pharmacia) to protect internal PstI sites from subsequent digestion. Double-stranded cDNA was then repaired with T4 DNA polymerase, ligated with HindIII/XmnI adaptors (New England Biolabs), digested with PstI, size selected to remove cDNAs smaller than 500 bp (15), and cloned into HindIII/PstI-digested pBluescript SK(+) (Stratagene). The ligated plasmid was transformed into XL-1 Blue MRF′ (Stratagene) to obtain a library of 5 × 105 independent cDNA clones.
The normalized RM cDNA library was prepared from single-stranded RM cDNA eluted from genomic DNA beads (see below). Single-stranded cDNA was converted to double-stranded cDNA using the pBluescript KS primer, cloned into pBluescript SK(+), and transformed into XL-1 Blue MRF′ as described above. A normalized library of 4.4 × 104 independent cDNA clones was obtained.
Preparation of Genomic DNA-Coated Magnetic Beads and Normalization of the RM cDNA Library.
Genomic Drosophila DNA was partially digested with Sau3A and MaeIII, size fractionated, and a Klenow “fill in” reaction (15) was used to incorporate biotin-dUTP (Enzo Biochem) into the ends of the Sau3A and MaeIII fragments. The biotin-labeled genomic DNA was immobilized on streptavidin-coated magnetic beads (Dynal, Greak Neck, NY) using a modification of the manufacturer’s instructions. The beads were collected, washed, and used immediately for cDNA hybridization.
To prepare single-stranded cDNA “driver” for hybridization to the genomic DNA “target,” the RM library was transcribed in vitro and the product RNA subsequently converted into single-stranded cDNA. The genomic DNA beads were resuspended in hybridization mix containing single-stranded RM cDNA as driver and free polysome poly(A)+ RNA as competitor to block the hybridization of free polysome cDNA to the beads. The beads were hybridized at 65°C for 16 hr with rocking. After hybridization the beads were washed extensively and subsequently the hybridized cDNA was eluted and recovered by ethanol precipitation. The protocol used to construct the library is shown schematically in Fig. 1.
Figure 1.
Schematic representation of the cDNA normalization procedure. The normalization method is described in detail in the text.
Whole-Mount RNA in Situ Hybridization of Drosophila Embryos in 96-Well Plates.
The nonradioactive whole-embryo in situ hybridization method described by Tautz and Pfeifle (16) was adapted to the use of RNA probes to achieve maximum sensitivity. To allow expedient screening with large numbers of probes, the protocol was further modified for hybridization in 96-well plates. Staging of embryos and description of expression domains were performed as described (17) using a standardized vocabulary (http://flybase.bio.indiana.edu/docs/flydocs/flybase/controlled-vocabularies.txt).
Photography and Digital Imaging.
Between 10 and 15 individually staged embryos were selected for photography for each RM cDNA clone. Expression domains were examined using Nomarski optics on an Axiophot microscope (Zeiss) and photographed using standard 35-mm film. Digital images were generated and written onto compact discs (Eastman Kodak).
DNA Sequencing and Analysis.
The cDNAs were sequenced using either the ABI Prism Dye Terminator Cycle Sequencing Ready Reaction kit or the Pharmacia Autoread Sequencing kit and the products were run on an ABI Prism 373 DNA Sequencer or a Pharmacia ALF Express DNA Sequencer, respectively. The resulting DNA sequences were trimmed and edited using Sequencher 3.1 software. Edited sequences average about 350–400 nts in length and contain 3% or less ambiguity. In cases where sequences from the 5′ and 3′ ends of the insert overlapped, contigs were constructed. Database searches were carried out using the BLASTN and TBLASTX programs (18).
Database and Software.
We implemented the cDNA database in Illustra version 3.2, an object-oriented relational database. The network browser interface was supported by the Apache v1.2.5 HTTP server. Common Gateway Interface scripts were written in Perl v1.0.5. Assemblies of the cDNA sequences are publicly viewable using a Java applet. The applet was compiled with Java 1.0.3 and utilized the BDGP/Neomorphic Software Inc. widget set. The cDNA sequences were analyzed using gapped WU-BLAST v2.0 (Warren Gish). Consensus sequences from multiple cDNAs (tentatively the same gene) were assembled using PHRAP (P. Green, in preparation).
RESULTS
Isolation of mRNA from RMs.
Most mRNAs that encode membrane and secreted proteins are bound to the rough endoplasmic reticulum through ribosomes engaged in cotranslational secretion of their nascent polypeptides. We isolated rough endoplasmic reticulum membranes, or RMs, from embryos as a source of mRNAs encoding membrane and secreted proteins. We found that only a small fraction of polysomal mRNA (<10%) is present in the RM preparation; the vast majority of embryonic mRNA appears to be translated on “free” polysomes encoding cytosolic proteins. This result is consistent with sequencing data obtained from an embryo cDNA library prepared from unfractionated mRNA, which revealed that 94% of clones with matches to known proteins encoded intracellularly localized proteins (see below).
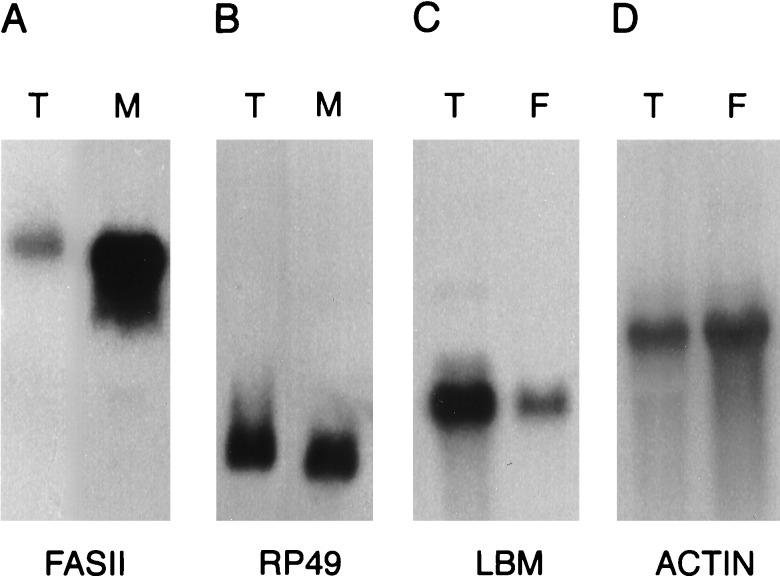
Northern blot analysis was used to determine the extent to which mRNAs encoding membrane and secreted proteins are enriched in the RM RNA preparation (Fig. 2 A and B). The results show that the mRNA encoding the membrane protein fasciclin II (Fas II) is approximately 10-fold enriched in the RM RNA preparation relative to the mRNA encoding the cytosolic protein rp 49. Similar results were obtained using probes representing other membrane and cytosolic proteins (data not shown). Although these results confirm that the RM RNA preparation is enriched for mRNAs encoding membrane and secreted proteins, they also reveal that the RM preparation was contaminated with significant amounts of free polysomes. The low yield of RMs obtained from embryos and the RNA degradation suffered on sucrose gradients precluded further purification of the RM preparation.
Figure 2.
mRNAs encoding transmembrane proteins are selectively enriched in the Rm RNA fraction and decreased in the free polysome fraction. (A and B) Northern blots containing 20 μg of RNA from the total (T) or RM (M) fractions were hybridized with the genes encoding the transmembrane protein Fas II (A) (4,500 nt transcript) or the rp 49 ribosomal protein (B) (600 nt transcript). (C and D) Northern blots containing 10 μg of poly(A)+ RNA from the total (T) or free polysome (F) fractions were hybridized with genes encoding the transmembrane protein late bloomer Lbm (C) (1,300 nt transcript) or the cytosolic protein actin 57B (D) (2,000 nt transcript).
Preparation of a Normalized cDNA library.
Poly(A)+ RNA was prepared from RM RNA and used to generate a directionally cloned RM cDNA library (see Materials and Methods). To increase the chances of identifying genes that encode low abundance mRNAs, it was important to normalize the representation of cDNAs in this library. A method of normalization was needed that would increase the prevalence of rare cDNAs encoding membrane and secreted proteins without increasing the prevalence of cDNAs encoding cytosolic proteins. The normalization procedure we developed is based on hybridizing a large excess of single-stranded cDNA to a limiting amount of genomic DNA that is attached to magnetic beads (Fig. 1). To prevent cDNAs encoding cytosolic proteins from hybridizing to the genomic DNA-coated beads, free polysome poly(A)+ RNA was added as a competitor. Once the hybridization was complete, the unbound cDNA was discarded and the normalized library was prepared from the cDNA that hybridized to the genomic DNA. Thus, the representation of cDNAs in the normalized library should reflect gene copy number, rather than mRNA abundance.
The effectiveness of this method was determined by colony blot hybridization using probes to a moderately abundant RM-bound mRNA (Fas II), a low abundance RM-bound mRNA (connectin,) and a cytosolic mRNA (Ras 1). As expected, normalization had the greatest effect on the frequency of clones representing the low abundance connectin mRNA, which showed a 13-fold increase from an initial frequency of 1 in 90,000 clones to 1 in 6,900. By comparison, the frequency of Fas II clones in the normalized library increased only 2-fold from an initial frequency of 1 in 10,000 clones to 1 in 4,300. Unexpectedly, the frequency of Ras 1 clones in the library also increased substantially (6-fold from an initial frequency of 1 in 130,000 clones to 1 in 21,000). This suggests that the addition of free polysome RNA as a competitor in the hybridization mix was only partially effective at preventing normalization of cDNAs encoding cytosolic proteins. Given that typical embryo cDNA libraries contain similar numbers of Fas II and Ras 1 clones (data not shown), the results suggest that the normalized RM cDNA library is approximately 5-fold enriched for clones encoding membrane and secreted proteins.
Since normalization of the RM library resulted in an increase in the representation of cDNAs encoding cytosolic proteins, we devised a rapid Northern blot assay to determine whether a cDNA of interest is likely to encode a membrane or secreted protein or a cytosolic protein (Fig. 2 C and D). Specifically, the cDNA is hybridized to a blot containing one lane of unfractionated mRNA and one lane of free polysome mRNA: if the hybridization signal is decreased in the free polysome lane, this suggests that the mRNA was bound to RMs and thus encodes a membrane or secreted protein. To date, this assay has produced accurate predictions for 11 of 12 cDNAs tested (data not shown).
RNA in Situ Hybridization of cDNA Clones to Drosophila Embryos.
Spatial and temporal embryonic expression profiles of the genes represented by RM cDNAs were determined by RNA in situ hybridization to whole-mount Drosophila embryos. To evaluate large numbers of cDNA probes, we developed an RNA in situ hybridization protocol that allows the simultaneous screening of 96 different RNA probes in a single multiwell plate.
A total of 2,518 RNA probes prepared from individual, randomly picked cDNA clones was screened on 0- to 24-hr old whole-mount embryos. Of these clones, 917 (36%) were expressed in specific patterns during embryogenesis, whereas 1,206 (48%) of the cDNAs showed apparent uniform expression throughout the embryo. The remaining 395 clones (16%) did not produce detectable levels of staining in the embryo. For every cDNA clone with specific expression patterns, 10–15 embryos covering a range of different embryological stages (starting at the fertilized egg to stage 16) were evaluated and photographed. As expected, a wide variety of temporal and spatial expression patterns were observed (examples in Fig. 3).
Figure 3.

Expression domains of a subset of RM clones. The RNA expression patterns of selected RM clones in distinct parts of the Drosophila embryo are shown. A typical image assigned to each RM clone in the database is shown in A, while B through L show a detail of these images. In B through L, anterior is to the left. (A) Expression of CK02213 in the anterior and posterior midgut primordium (arrows), the midgut (arrowhead), and the visceral mesoderm. This clone shows homology to the human NMDA receptor glutamate-binding subunit. (B) Expression of CK02262 in the ventral nerve cord and brain. This clone shows homology to the Bos taurus gene for Na/Ca,K exchanger protein. (C) Expression of CK02467 in the proventriculus, a part of the stomodeum. This clone does not show homology to any genes in the existing gene databases. (D) Expression of CK01670 in the developing tracheal system. This clone does not show homology to any genes in the existing gene databases. (E) Expression of CK01209 in the brain. This clone shows homology to human serine/threonine kinase. (F) Expression of CK02623 in the salivary glands and proventriculus. This clone shows homology to the rat Na2+-dependent inorganic phosphate cotransporter. (G) Expression of CK00246 in the central nervous system, ventral nerve cord, and brain. This clone shows homology to mouse and human ESTs. (H) Expression of CK01174 in the reproductive system (gonads). This clone does not show homology to any genes in the existing gene databases. (I) Expression of CK00490 in the anterior and posterior midgut primordium. This clone shows homology to several human ESTs. (J) Expression of CK01593 in the dorsal vessel and lymph gland. This clone does not show homology to any genes in the existing gene databases. (K) Expression of CK02229 in the epidermis, visceral mesoderm, tracheal system, and fore and hindgut. This clone shows homology to human laminin. (L) Uniform expression of CK02318 throughout the epidermis. This clone shows homology to a C. elegans EST.
The frequency with which cDNAs were found to be expressed in various embryonic organs is summarized in Table 1 (ubiquitously expressed cDNAs are not included). The numbers shown in Table 1 are adjusted for multiple occurrences of cDNAs representing a single gene. A disproportionately large number of cDNAs are expressed in the embryonic gut, the central nervous system, and the muscle, whereas only a small percentage of cDNAs are found in tissues such as the amnioserosa, glands, trachea, imaginal discs, and gonads. A possible explanation for this observation is that expression in a tissue such as the gut is more easily scored than, for example, that in the embryonic imaginal discs; these consist of only 10–25 cells and are considerably more difficult to identify.
Table 1.
Expression domains of RM clones during embryogenesis
| Spatial expression domain | RM clones*, n | %‡ |
|---|---|---|
| Fertilized egg | 167 (282) | 7 |
| Blastoderm | 13 (18) | <1 |
| Gastrula | 9 (9) | <1 |
| Segmented germ band | 4 (5) | <1 |
| Epidermis | 86 (134) | 4 |
| Mesoderm | 379 (638) | 16 |
| Somatic mesoderm | 87 (160) | 4 |
| Visceral mesoderm | 228 (329) | 9 |
| Head mesoderm | 28 (84) | 1 |
| Muscle | 36 (65) | 2 |
| Nervous system | 210 (317) | 9 |
| Stomatogastric nervous system | 6 (8) | <1 |
| Peripheral nervous system | 13 (27) | <1 |
| Central nervous system | 191 (282) | 8 |
| Embryonic gut | 418 (642) | 17 |
| Foregut | 99 (129) | 4 |
| Midgut | 169 (284) | 7 |
| Hindgut | 94 (136) | 4 |
| Malpighian tubule | 38 (72) | 2 |
| Gastric caecum | 18 (21) | <1 |
| Amnioserosa | 28 (41) | 1 |
| Embryonic glands | 69 (95) | 3 |
| Embryonic tracheal system | 25 (32) | 1 |
| Reproductive system | 24 (43) | 1 |
| Imaginal disc | 3 (6) | <1 |
The first number given is the number of cDNAs that represent unique sequences, and the number in parentheses is the total number of clones. Individual clones are usually expressed in more than one tissue. Uniformly expressed cDNAs are not included.
The percentage of unique clones in the database expressed in a particular tissue.
Only a small percentage of the clones were found to be expressed during early zygotic stages of development (blastoderm, gastrula, and segmented germ band stages). The vast majority are expressed during stages when the internal organs, like the gut, the central nervous system, and the muscles are formed. Because the embryos that were used to make the cDNA library were taken from an 8- to 16-hr collection, the period when these tissues are developing, the bias toward cDNAs expressed in the internal organs is not unexpected. In addition, a large number of cDNAs show hybridization to early-stage embryos prior to the onset of zygotic gene expression. This hybridization presumably represents maternal contribution of the cognate mRNAs.
Sequence Analysis.
We next set out to sequence the 5′ and 3′ ends of the 917 cDNAs that represent genes with tissue- and stage-specific expression patterns, since these genes are good candidates to play important roles in development. In addition, we sequenced a subset (381) of the cDNAs that represent uniformly expressed genes. Based on sequence analysis, we were able to identify 297 recurring cDNAs. The largest class of repetitive cDNAs corresponded to mitochondrial genes, which we found to be strongly expressed in the visceral mesoderm. The relatively high prevalence of mitochondrial cDNAs is likely due to the fact that mitochondria are a significant contaminant of RM preparations and mitochodrial DNA is present at a very high copy number in embryos. After taking redundancies into account, the 1,298 sequenced cDNAs represent 1,001 unique sequences. This is likely to be a slight overestimate of the number of different genes represented, however, since a single gene can produce transcripts with different 3′ ends and “false” 3′ ends can be generated by internal priming during cDNA synthesis. Thus, we expect the number of different genes examined to be between 800 and 900.
This sequence data provided us with another opportunity to assess the enrichment of the library for cDNAs encoding membrane-targeted proteins. Of the 1,001 different sequences, 124 correspond to known Drosophila genes for which we could predict a subcellular localization based on protein similarity or published protein localization data; 47 of these genes encode membrane proteins and 77 encode either nuclear or cytoplasmic proteins. Thus, approximately 38% of the cDNAs that correspond to known genes encode for membrane proteins. For comparison, we carried out a similar analysis on sequences from an unfractionated embryonic cDNA library, the LD library (sequence data made available by the Berkeley Drosophila Genome Project; http://fruitfly.berkeley.edu). We analyzed 326 LD cDNAs that correspond to known Drosophila genes. These cDNAs represent 147 different genes, of which 16 (11%) encode membrane proteins and 131 (89%) encode nuclear or cytoplasmic proteins. These results suggest that the RM library is approximately 3.5-fold enriched for cDNAs encoding membrane-targeted proteins, similar to the 5-fold enrichment suggested by our colony blot hybridization results (discussed above). It should be noted that sequence analysis may underestimate the overall representation of clones encoding membrane-targeted proteins in the RM library due to a bias for cytosolic and nuclear proteins in the Drosophila sequence database. To date, six of eight RM cDNAs characterized solely on the basis of expression pattern have been found to encode membrane or secreted proteins (data not shown).
The 811 sequences that did not correspond to previously described Drosophila genes were analyzed for homology to translated nucleotide databases using the TBLASTN program (18). We found that 267 of these sequences show significant similarity to characterized genes in other species (i.e., homologies that have a probability of 10−5 or less and that are not the result of simple repetitive sequences). As expected, many of these cDNAs encode for homologs of mammalian membrane and secreted proteins, including growth factors, transmembrane receptors, ion transporters, and proteins that function in the endoplasmic reticulum (Table 2). Another 125 sequences show significant homology to identified but uncharacterized sequences in other organisms, typically to human and mouse expressed sequence tags (ESTs) and to Caenorhabditis elegans genomic DNA. The remaining 419 sequences have no significant homology to any sequence in the databases. Since the majority of the cDNAs are relatively small (approximately 1 kb), it is likely that many of the sequences consist mainly of 3′ untranslated region and therefore would not be useful for searching databases for protein homologies. Therefore, the percentage of Drosophila genes that have homologs in other species is likely to be significantly higher than these results suggest.
Table 2.
Selected RM cDNAs with homologies to known mammalian genes
| Clone no. | Highly similar mammalian gene |
|---|---|
| CK02126 | Human epidermal surface antigen (M60922) |
| CK02288 | Human plasma membrane calcium ATPase isoform 3x/b (U60414) |
| CK01423 | Human stomatin (X60067) |
| CK01140 | Human adenosine triphosphate (M95541) |
| CK00230 | Human KDEL receptor (X55885) |
| CK00459 | Rat purine-specific Na+ nucleoside cotransporter (U25055) |
| CK01227 | Human multidrug resistance associated protein (L05628) |
| CK02656 | Mouse ABC8 (Z48745) |
| CK00309 | Canine docking protein (SRP receptor) (X06272) |
| CK01110 | Human testican (X73608) |
| CK00043 | Human SEC13R membrane protein (L09260) |
| CK00325 | Human sulfonylurea receptor (L40625) |
| CK01510 | Human K-Cl cotransporter, hKCC1 (U55054) |
| CK01296 | Rat TRAP complex γ subunit (Z14030) |
| CK02248 | Rat Dri42 (Y07783) |
| CK01027 | Human bumetanide-sensitive Na-K-Cl cotransporter (U30246) |
| CK02682 | Mouse reticulocalbin (D13003) |
| CK00198 | Mouse macrophage scavenger receptor (M59445) |
| CK01823 | Human E16 (M80244) |
| CK00539 | Human LDL-receptor related protein (X13916) |
| CK01577 | Mouse scavenger receptor class B type I (mSR-BI) (U37799) |
| CK02137 | Rat zinc transporter, ZnT-2 (U50927) |
| CK02567 | Mouse thrombospondin, THBS2 (M64866) |
These clone-gene combinations show TBLASTX values between e−18 and e−59. For each mammalian gene, the GenBank accession number is shown in parentheses.
Data Availability over the Internet.
A database describing the expression patterns and DNA sequences of the cDNAs compiled in this study that were expressed in specific tissues is accessible at http://fruitfly.berkeley.edu. The web page describing each EST shows the sequence, accession numbers, and a summary of gene expression data along with a low-resolution expression image and a summary of similarity to other sequences. A high-resolution digital image is available for downloading. Several types of searches are available to query this information: (i) Expression Domain Keyword Search: Every expression image has been annotated using the standardized set of terms developed by Flybase for the description of Drosophila anatomy (http://flybase.bio.indiana.edu). Therefore, keyword searches for cDNAs that are expressed in a particular embryonic organ, or combination of organs, may be performed; (ii) Sequence Keyword Search: A BLAST similarity search was performed on each EST and the results stored in the database, including the accession number of the GenBank entries of similar sequences. cDNAs that show similarity to a particular class of gene may be found by searching for words or phrases that are likely to be found in the gene’s GenBank description; (iii) Clone Identifier Search: unique identifiers, such as the clone name (CK number) or accession number, can be used to retrieve an individual cDNA record; (iv) Sequence Similarity Search: Using a public BLAST server available at the same site as the database, searches for ESTs similar to any query sequence can be performed.
DISCUSSION
We have used high throughput whole-embryo in situ hybridization and a normalized cDNA library prepared from RM-bound mRNA to identify membrane and secreted proteins whose expression is associated with specific developmental processes during embryogenesis. The expression patterns of 1,003 individual cDNAs and sequence information for 1,298 cDNAs are available on a public database (http://fruitfly.berkeley.edu). This database makes it possible to rapidly identify new developmentally regulated genes and, based on the sequence and expression pattern, formulate testable hypotheses for the function of the genes. For example, based on a motoneuron-specific expression pattern in the developing nerve cord, we identified the first Drosophila member of the tetraspanin family of transmembrane proteins, Late Bloomer (19). Through subsequent genetic analysis, we determined that late bloomer function facilitates neuromuscular synapse formation in the embryo (19). Similarly, characterization of a cDNA expressed specifically in muscle led to the identification of a new Drosophila glutamate receptor (20).
Although the RM cDNA library is 4- to 5-fold enriched for membrane and secreted proteins, this library also contains a large fraction of cDNAs encoding cytosolic and nuclear proteins. This is due in part to the fact that embryonic mRNAs encoding membrane and secreted proteins appear to be much less abundant than mRNAs encoding cytosolic and nuclear proteins. In addition, normalization of the RM library decreased the enrichment for membrane and secreted proteins by partially restoring the prevalence of clones encoding cytosolic and nuclear proteins. In spite of this drawback to normalization, we chose to screen the normalized RM cDNA library to reduce the number of recurrent cDNAs and thereby increase the chances of identifying less abundant mRNAs whose expression is limited to a small number of cells in the embryo.
The normalization method we describe has both advantages and disadvantages relative to the more standard methods of normalizing by limited cDNA self-hybridization (21). The main advantage of normalizing by hybridization to genomic DNA is that the method requires no optimization of hybridization times or titration of hydroxyapatite elution conditions. However, genomic DNA hybridization normalizes on the basis of gene copy number, which means that high copy number genes are overrepresented in the cDNA library. We found mitochondrial genes were particularly problematic; approximately 15% of the clones in the library represent mitochondrial genes. This could be resolved by further purification of the genomic DNA to ensure that mitochodrial DNA is not present on the magnetic beads. Another limitation of the technique is the need for relatively large amounts of genomic DNA target in the hybridization to capture enough cDNA to prepare a library. The amount of DNA needed for genomes of higher complexity than Drosophila would necessitate a much larger amount of genomic DNA-coated beads, which would increase the amount of contamination in the library due to nonspecific hybridization. Also, the larger amount of interspersed repetitive DNA in vertebrate genomes would cause rapid annealing of the genomic DNA and could cause vast overrepresentation of mRNAs containing repetitive elements in their untranslated regions. For these reasons, this normalization technique may not be appropriate for vertebrate genomes.
Subcellular fractionation of RM-bound mRNA is a convenient way to prepare mRNA enriched for membrane and secreted proteins. However, it requires a relatively large amount of tissue to isolate enough mRNA to generate a library that does not require amplification by PCR. It is also difficult to normalize a RM library without increasing the prevalence of mRNAs encoding cytosolic and nuclear proteins. In the course of this work, two alternative methods for identifying cDNAs encoding membrane and secreted proteins were described that have some advantages over subcellular fractionation (22, 23). These methods are based on transforming tissue culture cells (22) or yeast (23) with a vector that will express an assayable reporter protein only when a cDNA encoding a signal sequence is cloned into the vector. This approach allows cDNA libraries to be prepared from small amounts of unfractionated mRNA, and the library of positive cDNAs that is generated is highly specific for membrane and secreted proteins.
The Drosophila genome is estimated to contain approximately 12,000 genes (5). The fact that we were able to carry out in situ hybridization to embryos for over 2,500 different cDNA clones in this study argues that the methodology we describe could be used to collect similar data for all Drosophila genes. Suitable probes could be derived by using PCR to amplify segments of sequenced genomic DNA or cDNA clones as templates. The highly sensitive and rapid in situ hybridization method used here allows the detailed visualization of gene expression and provides a level of spatial and temporal resolution that is not currently obtainable by methods that require RNA isolation and hybridization to clone (24) or oligonucleotide (25) arrays. Such expression data, along with the more quantitative data provided by hybridization to arrays, will be essential for deciphering gene regulatory networks.
Acknowledgments
We thank Fred Wolf for his help with the initial RNA in situ screens, Rick Fetter and Lee Fradkin for helping to prepare the figures, and Lee Fradkin and the members of the Rubin and Goodman laboratories for critical review of the manuscript. C.C.K. was supported as a Jane Coffin Childs postdoctoral fellow and a Howard Hughes Medical Institute postdoctoral associate. T.L.S. is a Jane Coffin Childs postdoctoral fellow. J.N.N. is a postdoctoral associate and C.S.G. and G.M.R. are investigators with the Howard Hughes Medical Institute. This work was supported in part by National Institutes of Health Grant HG00750.
ABBREVIATIONS
- RM
rough microsome
- EST
expressed sequence tag.
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AA140639–AA140951; AA140953–AA140968; AA140971–AA141066; AA141069–AA141114; AA141116–AA141344; AA141346–AA141693; AA141695–AA141908; AA141911–AA141987; AA141989–AA142102; AA142105–AA142202; AA142205–AA142210; AA142213–AA142231; AA142233–AA142250; AA142252–AA142269; AA142271–AA142312).
A commentary on this article begins on page 9716.
References
- 1.Burke R, Basler K. Curr Opin Neurobiol. 1997;7:55–61. doi: 10.1016/s0959-4388(97)80120-1. [DOI] [PubMed] [Google Scholar]
- 2.Perrimon N. Cell. 1996;86:513–516. doi: 10.1016/s0092-8674(00)80124-5. [DOI] [PubMed] [Google Scholar]
- 3.Derynck R, Zhang Y. Curr Biol. 1996;6:1226–1229. doi: 10.1016/s0960-9822(96)00702-6. [DOI] [PubMed] [Google Scholar]
- 4.Cavallo R, Rubenstein D, Peifer M. Curr Opin Genet Dev. 1997;7:459–466. doi: 10.1016/s0959-437x(97)80071-8. [DOI] [PubMed] [Google Scholar]
- 5.Miklos G L, Rubin G M. Cell. 1996;86:521–529. doi: 10.1016/s0092-8674(00)80126-9. [DOI] [PubMed] [Google Scholar]
- 6.Wilson C, Pearson R K, Bellen H J, O’Kane C J, Grossniklaus U, Gehring W J. Genes Dev. 1989;3:1301–1313. doi: 10.1101/gad.3.9.1301. [DOI] [PubMed] [Google Scholar]
- 7.Bier E, Vaessin H, Shepherd S, Lee K, McCall K, Barbel S, Ackerman L, Carretto R, Uemura T, Grell E, Jan L Y, Jan Y N. Genes Dev. 1989;3:1273–1287. doi: 10.1101/gad.3.9.1273. [DOI] [PubMed] [Google Scholar]
- 8.Torok T, Tick G, Alvarado M, Kiss I. Genetics. 1993;135:71–80. doi: 10.1093/genetics/135.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spradling A C, Stern D M, Kiss I, Roote J, Laverty T, Rubin G M. Proc Natl Acad Sci USA. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kidwell M G. In: Drosophila: A Practical Approach. Roberts E D, editor. Washington, DC: IRL; 1986. pp. 59–83. [Google Scholar]
- 11.Bastiani M J, Harrelson A L, Snow P M, Goodman C S. Cell. 1987;48:745–755. doi: 10.1016/0092-8674(87)90072-9. [DOI] [PubMed] [Google Scholar]
- 12.Zipursky S L, Venkatesh T R, Teplow D B, Benzer S. Cell. 1984;36:15–26. doi: 10.1016/0092-8674(84)90069-2. [DOI] [PubMed] [Google Scholar]
- 13.Gaetani S, Smith J, A, Feldman R A, Morimoto T. Methods Enzymol. 1983;96:3–24. doi: 10.1016/s0076-6879(83)96005-6. [DOI] [PubMed] [Google Scholar]
- 14.Natzle J E, Hammonds A S, Fristrom J W. J Biol Chem. 1986;261:5575–5583. [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 16.Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 17.Hartenstein V. Atlas of Drosophila Development. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. [Google Scholar]
- 18.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Kopczynski C C, Davis G W, Goodman C S. Science. 1996;271:1867–1870. doi: 10.1126/science.271.5257.1867. [DOI] [PubMed] [Google Scholar]
- 20.Petersen S A, Fetter R D, Noordermeer J N, Goodman C S, DiAntonio A. Neuron. 1997;19:1237–1248. doi: 10.1016/s0896-6273(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 21.de Fatima Bonaldo M, Lennon G, Soares M B. Genome Res. 1996;6:791–806. doi: 10.1101/gr.6.9.791. [DOI] [PubMed] [Google Scholar]
- 22.Tashiro K, Tada H, Heilker R, Shirozu M, Nakano T, Honjo T. Science. 1993;261:600–603. doi: 10.1126/science.8342023. [DOI] [PubMed] [Google Scholar]
- 23.Klein R D, Gu Q, Goddard A, Rosenthal A. Proc Natl Acad Sci USA. 1996;93:7108–7113. doi: 10.1073/pnas.93.14.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 25.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, et al. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
A database describing the expression patterns and DNA sequences of the cDNAs compiled in this study that were expressed in specific tissues is accessible at http://fruitfly.berkeley.edu. The web page describing each EST shows the sequence, accession numbers, and a summary of gene expression data along with a low-resolution expression image and a summary of similarity to other sequences. A high-resolution digital image is available for downloading. Several types of searches are available to query this information: (i) Expression Domain Keyword Search: Every expression image has been annotated using the standardized set of terms developed by Flybase for the description of Drosophila anatomy (http://flybase.bio.indiana.edu). Therefore, keyword searches for cDNAs that are expressed in a particular embryonic organ, or combination of organs, may be performed; (ii) Sequence Keyword Search: A BLAST similarity search was performed on each EST and the results stored in the database, including the accession number of the GenBank entries of similar sequences. cDNAs that show similarity to a particular class of gene may be found by searching for words or phrases that are likely to be found in the gene’s GenBank description; (iii) Clone Identifier Search: unique identifiers, such as the clone name (CK number) or accession number, can be used to retrieve an individual cDNA record; (iv) Sequence Similarity Search: Using a public BLAST server available at the same site as the database, searches for ESTs similar to any query sequence can be performed.