Abstract
The evolutionarily conserved mammalian Sin3 (mSin3) transcriptional corepressor interacts with a diverse array of transcription factors mainly through two PAH (paired amphipathic helix) domains located near the N terminus. Previous studies suggested the possibility of interdomain interactions involving the PAH domains. Here, we show that the domains are structurally independent and the properties of the individual domains, such as the conformational heterogeneity and the ability of mSin3A PAH2 to homodimerize, are preserved in constructs that span both PAH domains. Our results thus suggest that the N-terminal segments of the Sin3 proteins are broadly available for interactions with other proteins and that the PAH domains are organized into structurally independent modules. Our data also rule out any heterotypic association between the paralogous mSin3A and mSin3B proteins via interactions involving the mSin3A PAH2 domain.
Keywords: Sin3 corepressor, protein–protein interaction, PAH domain, NMR, transcription regulation
The recruitment of chromatin-modifying and/or chromatin-remodeling enzymes by sequence-specific DNA-binding factors is a common, albeit important, step in the pathway leading to the activation or repression of target gene transcription in eukaryotes (Ptashne and Gann 2002). An impressive array of chromatin-modifying and chromatin-remodeling activities has been identified over the past decade, but these enzymes are frequently found in large multiprotein coregulator complexes, with the additional subunits presumably lending specificity to the process (Elgin and Workman 2000). The Sin3 corepressor complex comprising at least 10 subunits is one of only a handful of major corepressor complexes identified thus far in mammalian cells that exerts its negative effects on gene transcription through the action of histone deacetylases (HDACs) (Hassig et al. 1997; Laherty et al. 1997, 1998; Zhang et al. 1998; Alland et al. 2002; Fleischer et al. 2003). The Sin3 corepressor has a dual role, functioning not only as a molecular scaffold for complex assembly but also as a molecular adapter, bridging HDACs with an astonishingly large and diverse group of DNA-binding transcription factors and chromatin-binding proteins (Ayer 1999; Knoepfler and Eisenman 1999; Silverstein and Ekwall 2005). The interactions are generally mediated through one or more of six discrete regions conserved from yeast to human including four imperfect copies of the PAH domain. The second copy of the PAH domain (PAH2) is the site for interactions with numerous factors including members of the Mad family (McArthur et al. 1998; Schreiber-Agus and DePinho 1998), Mnt/Rox (Hurlin et al. 1997), Pf1 (Yochum and Ayer 2001), HBP1 (Swanson et al. 2004), the KLF family (Zhang et al. 2001), TGIF (Wotton et al. 2001), Menin (Kim et al. 2003), MNF-β (Yang et al. 2000), and EKLF (Chen and Bieker 2004).
Our previous structural studies revealed that the mammalian Sin3A (mSin3A) PAH2 domain interacted with diverse targets through a hydrophobic cleft by binding to chain-reversible sequence motifs, thereby defining at least two distinct classes of PAH2 interactors (Brubaker et al. 2000; Swanson et al. 2004). The apo-mSin3A PAH2 domain exhibits conformational heterogeneity, with one of the conformers existing in a partially unfolded state, and homodimerizes with modest affinity, suggesting plausible mechanisms for binding to a broad range of targets through new surfaces on the one hand and for regulating Sin3 function via occlusion of the hydrophobic cleft on the other (Zhang et al. 2006). The PAH2 domain of the paralogous mSin3B protein engages targets in a manner similar to mSin3A PAH2 (Spronk et al. 2000; van Ingen et al. 2004), but the apo-protein is monomeric and is fully folded, although a small population of a minor conformer has been reported (van Ingen et al. 2006). The properties of these domains in the context of the full-length or much larger fragments of the Sin3 protein are unexplored, and the assumption of the autonomous nature of the individual domains at the structural level has not been thoroughly tested.
Results and Discussion
The weak tendency of the apo-mSin3A PAH2 domain to dimerize (Kd = 280 μM) prompted us to ask whether this might reflect an innate ability to interact with related PAH domains. Biologically, this could be a mechanism to thwart low-affinity interactions with the ubiquitous Φ-X-X-Φ-Φ sequence motifs found in transcription factors (Plevin et al. 2005). Potential candidates for mSin3A PAH2 interactions include the mSin3B PAH2 domain with which the domain shares 65% sequence identity and 81% similarity, the mSin3A PAH1 domain (33% identity, 63% similarity), and mSin3B PAH1 (36% identity, 56% similarity). The PAH3 and PAH4 domains were not considered as potential interactors as they are either constitutively associated with core components of the corepressor complex (e.g., SAP30 in the case of PAH3) (Laherty et al. 1998; Zhang et al. 1998) or have diverged substantially at the sequence level (e.g., PAH4) and are considered unrelated to the other PAH domains.
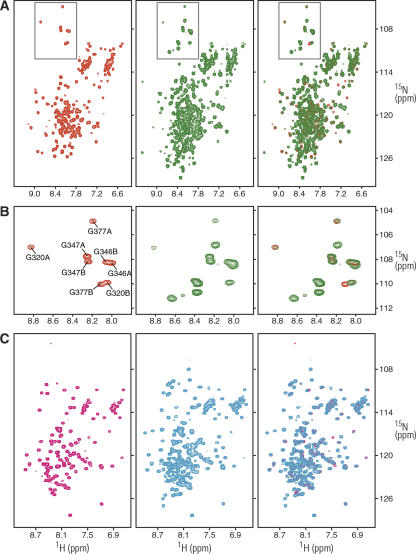
For the initial studies, we titrated purified mSin3A PAH1 with 15N-labeled mSin3A PAH2. We also expressed and purified mSin3B PAH2 and titrated with 15N-mSin3A PAH2. At submillimolar concentrations, neither mSin3A PAH1 nor mSin3B PAH2 produced any changes in the 1H-15N heteronuclear single quantum coherence (HSQC) spectra of mSin3A PAH2 (data not shown). Since weak intermolecular association could translate to a stronger intramolecular interaction when the domains are tethered in cis, we generated constructs spanning the PAH1 and PAH2 domains (designated PAH1/PAH2) of mSin3A. Expressing and purifying sufficient quantities of mSin3A PAH1/PAH2 was a challenge as the two domains are separated by a long linker region (∼110 residues) that rendered the protein readily susceptible to degradation by bacterial proteases. By optimizing the expression tags, inducing protein expression at low temperatures, and taking precautions to limit protease action during the purification stages, we were able to generate sufficient amounts of pure protein for NMR studies. Before dissolving the dry, lyophilized powder in NMR buffer, the integrity of the sample was confirmed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). NMR spectra were recorded immediately after sample preparation. The 1H-15N HSQC spectrum of this construct recorded at 0.35 mM concentration is characterized by a broad range of peak intensities and relatively modest amide proton chemical shift dispersion. The vast majority of (mostly intense) resonances fall between 7.5 and 8.5 ppm, typical of natively unfolded regions, most likely from the segment linking the two domains (Fig. 1A). The spectrum also shares similarities with the spectrum for the apo-PAH2 domain recorded at the same concentration under identical conditions (Zhang et al. 2006). Indeed, the conformational heterogeneity of apo-PAH2, which is characterized by the appearance of more than one set of correlations, is also evident in the apo-mSin3A PAH1/PAH2 spectrum (Fig. 1A). This is readily apparent in the upfield region of the 15N spectrum populated by glycine resonances (Fig. 1B). Seven out of eight correlations belonging to the four glycine residues in the PAH2 domain exhibit identical 1H and 15N shifts in the two constructs. The chemical shifts of the correlations belonging to Gly377 in the B-conformation differ slightly, although its origin is unknown and it might arise from subtle differences in solution conditions and/or sample concentrations. An overwhelming majority of the remaining correlations in the apo-mSin3A PAH2 spectrum overlaps with those in the apo-mSin3A PAH1/PAH2 spectrum (Fig. 1A, right panel). Some of the weaker correlations in the apo-mSin3A PAH2 spectrum (mainly those belonging to the B-conformation, whose population increases with concentration) (Zhang et al. 2006) exhibit diminished intensity in the apo-mSin3A PAH1/PAH2 spectrum, but this could be attributed in part to the substantial increase in overall molecular weight of the dimeric forms (∼60 kDa for apo-mSin3A PAH1/PAH2 vs. ∼20 kDa for apo-mSin3A PAH2). Furthermore, the PAH2 correlations in the PAH1/PAH2 spectrum exhibit similar concentration-dependent changes in chemical shifts noted previously in the apo-mSin3A PAH2 spectrum (Zhang et al. 2006) (data not shown). As in the case of the apo-PAH2, these changes likely reflect homodimerization of apo-PAH1/PAH2 mediated by PAH2, with the dimer exhibiting fast dissociation kinetics on the NMR timescale. Since our model for an intramolecular interaction between the PAH1 and PAH2 domains involves the same PAH2 surface that is used for homodimerization and interaction with other targets, a key prediction of this model would be the loss of conformational heterogeneity and homodimerization ability of the mSin3A PAH2 domain in the PAH1/PAH2 construct. The conservation of these mSin3A PAH2 properties in the apo-mSin3A PAH1/PAH2 construct thus strongly argues against this model.
Figure 1.
NMR spectra of the N-terminal PAH domains of mSin3A and mSin3B reveal the absence of intramolecular interactions between the domains. (A) 1H-15N correlated spectra of the mSin3A PAH2 (red), mSin3A PAH1/PAH2 (green), and an overlay of the two spectra (right) recorded at 25°C in 20 mM sodium phosphate buffer (pH 6.0). Protein concentrations were 0.35 mM, and identical NMR data acquisition, processing, display, and contouring threshold parameters were used. (B) Expanded plots corresponding to the glycine region (gray boxes) of the spectra shown in panel A. The assignments for the glycine residues in the two conformers (designated A and B) are shown (Zhang et al. 2006). (C) 1H-15N correlated spectra of mSin3B PAH2 (magenta), mSin3B PAH1/PAH2 (cyan), and an overlay of the two spectra (right) recorded at 25°C in 20 mM sodium phosphate buffer (pH 6.0). Protein concentrations were 0.24 mM, and identical NMR data acquisition, processing, display, and contouring threshold parameters were used.
In parallel with the studies described above, we also sought to explore possible interaction between the mSin3B PAH1 and PAH2 domains. A construct spanning the PAH1 and PAH2 domains of mSin3B was generated. The PAH domains of mSin3B, unlike its mSin3A counterpart, are separated by a much shorter linker region (∼50 residues). The integrity of the purified sample prior to NMR studies was again confirmed by SDS-PAGE. The quality of the 1H-15N HSQC spectrum of mSin3B PAH1/PAH2 is superior to that of a comparable construct of mSin3A (Fig. 1C). Intense correlations are seen ∼8 ppm in the proton spectrum, presumably from the natively unfolded linker segment. In contrast, the mSin3B 1H-15N HSQC spectrum is characterized by better dispersion with the hallmarks of a folded domain (van Ingen et al. 2006). Most of the correlations in the mSin3B PAH2 spectrum overlap well with those in the mSin3B PAH1/PAH2 spectrum (Fig. 1C). Additionally, no severe line-broadening effects are witnessed for PAH2 resonances in the PAH1/PAH2 spectrum, consistent with the absence of stable, intramolecular interactions involving the PAH domains. We note that two isoforms of mSin3B comprising the PAH1 and PAH2 domains have been described (Koipally et al. 1999). Our studies suggest that these isoforms are likely to antagonize the functions of the full-length forms by effectively competing for recruitment by cognate targets. Finally, titrations of unlabeled mSin3B PAH1/PAH2 with 15N-labeled mSin3A PAH2 at submillimolar concentrations produced no changes in the NMR spectrum (data not shown), ruling out heterotypic interactions between these segments.
In summary, we have tested whether the PAH2 domains of mSin3 proteins serve as interaction sites for related PAH domains. Our studies confirm the absence of such interdomain interactions at the inter- and intramolecular levels and suggest that the PAH1 and PAH2 domains of mSin3A and mSin3B are structurally independent. The N-terminal segments spanning the PAH1 and PAH2 domains within the respective proteins are thus likely to be broadly available for heterotypic protein–protein interactions. The weak propensity of the apo-mSin3A PAH2 domain to homodimerize and exist in two different conformations is preserved in the mSin3A PAH1/PAH2 construct. The properties of the apo-mSin3A PAH2 domain thus distinguish this domain from the other PAH1 and PAH2 domains.
Materials and Methods
Expression and purification of mSin3A PAH2, mSin3A PAH1/PAH2, mSin3B PAH2, and mSin3B PAH1/PAH2
The coding sequence of mSin3A PAH1/PAH2 corresponding residues 118–385 was amplified by PCR and inserted into the pMCSG7 expression vector (Stols et al. 2002). All cloned gene segments were confirmed by DNA sequencing. Escherichia coli BL21(DE3) cells (Novagen) containing the vector were grown at 37°C in M9 minimal media. The growth temperature was shifted to 20°C when the OD600 nm reached ∼0.7. Expression of the His6-tagged protein was induced using 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG), and the cells were harvested 16 h thereafter. Cell pellets were suspended in 20 mM sodium phosphate buffer (pH 7.5) containing 0.3 M sodium chloride, 2 mM Tris (2-carboxy-ethyl) phosphine hydrochloride (TCEP), 1 mM phenyl-methylsulfonyl (PMSF), 1 μM leupeptin, 1 mM pepstatin, and 0.1% Triton X-100, lysed via sonication, and centrifuged. The supernatant was incubated with the His-Select Nickel resin (Sigma) for 30 min. Bound proteins were cleaved from the resin by incubating with tobacco etch virus (TEV) protease for 4 h at 22°C followed by overnight incubation at 4°C. The mixture was centrifuged, and the desired protein in the supernatant was purified to homogeneity via reversed-phase HPLC using a C18 column (Vydac) and a linear gradient of 0.1% trifluoroacetic acid (TFA) and 0.1% TFA in 80% acetonitrile and lyophilized. An mSin3A PAH1/PAH2 sample uniformly labeled with 15N isotope was produced as described above, except that cells were grown in M9 minimal media containing 15N-ammonium sulfate (Spectra Stable Isotopes).
The coding sequences of mSin3A PAH2, mSin3B PAH2, and mSin3B PAH1/PAH2 domains corresponding to residues 295–385, 145–252, and 29–252, respectively, were amplified by PCR and inserted into the pMCSG10 expression vector (Stols et al. 2002). All cloned gene segments were confirmed by DNA sequencing. E. coli BL21(DE3) cells (Novagen) containing the vector were grown at 37°C in M9 minimal media. The growth temperature was shifted to 20°C when the OD600 nm reached ∼0.7. Expression of the GST-tagged protein was induced using 1 mM IPTG, and the cells were harvested 16 h thereafter. Cell pellets were suspended in PBS buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 8.5), 5 mM EDTA, 2 mM DTT, 1 mM PMSF, 1 μM leupeptin, 1 mM pepstatin, and 0.1% Triton X-100, lysed via sonication, and centrifuged. The supernatant was incubated with glutathione sepharose resin (GE Healthcare) for 30 min. Bound proteins were cleaved from the resin by incubating with TEV protease for 4 h at 22°C followed by overnight incubation at 4°C. The mixture was centrifuged, and the target protein in the supernatant was purified to homogeneity via reversed-phase HPLC using a C18 column and a linear gradient of 0.1% TFA and 0.1% TFA in 80% acetonitrile and lyophilized. Samples of mSin3A PAH2, mSin3B PAH2, and mSin3B PAH1/PAH2 uniformly labeled with 15N isotope were produced as described above, except that cells were grown in M9 minimal media containing 15N-ammonium sulfate.
NMR spectroscopy
NMR samples were prepared by dissolving the dry, lyophilized protein powder in 20 mM sodium phosphate buffer (pH 6.0) containing 0.2% (w/v) NaN3 and 2 mM DTT-d10. Protein concentrations were determined spectrophotometrically (Gill and von Hippel 1989). Identical sample concentrations were employed for recording NMR spectra of the recombinant mSin3A/B PAH2 and PAH1/PAH2 proteins. NMR data were acquired on a Varian Inova 600 MHz spectrometer at 25°C. NMR data processing and analysis were performed using an in-house modified version of Felix 98.0 (Felix NMR).
Acknowledgments
We thank Rebecca Imhoff and Richard Kang for technical assistance during the initial stages of this work. This work was supported by a grant from the NIH (GM 64715) to I.R. I.R. is a Scholar of the Leukemia and Lymphoma Society. Access to instrumentation in the WCAS Structural Biology NMR Facility at Northwestern University is gratefully acknowledged.
Footnotes
Reprint requests to: Ishwar Radhakrishnan, Northwestern University, Biochemistry, Molecular Biology, and Cell Biology Division, 2205 Tech Drive, Evanston, IL 60208-3500, USA; e-mail: i-radhakrishnan@northwestern.edu; fax: (847) 467-6489.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073097308.
References
- Alland, L., David, G., Shen-Li, H., Potes, J., Muhle, R., Lee, H.C., Hou H., Jr, Chen, K., DePinho, R.A. Identification of mammalian Sds3 as an integral component of the Sin3/histone deacetylase corepressor complex. Mol. Cell. Biol. 2002;22:2743–2750. doi: 10.1128/MCB.22.8.2743-2750.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayer, D.E. Histone deacetylases: Transcriptional repression with SINers and NuRDs. Trends Cell Biol. 1999;9:193–198. doi: 10.1016/s0962-8924(99)01536-6. [DOI] [PubMed] [Google Scholar]
- Brubaker, K., Cowley, S.M., Huang, K., Loo, L., Yochum, G.S., Ayer, D.E., Eisenman, R.N., Radhakrishnan, I. Solution structure of the interacting domains of the Mad–Sin3 complex: Implications for recruitment of a chromatin-modifying complex. Cell. 2000;103:655–665. doi: 10.1016/s0092-8674(00)00168-9. [DOI] [PubMed] [Google Scholar]
- Chen, X., Bieker, J.J. Stage-specific repression by the EKLF transcriptional activator. Mol. Cell. Biol. 2004;24:10416–10424. doi: 10.1128/MCB.24.23.10416-10424.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin, S.C.R., Workman, J.L. Chromatin structure and gene expression. 2d ed. Oxford University Press; New York: 2000. p. 328. [Google Scholar]
- Fleischer, T.C., Yun, U.J., Ayer, D.E. Identification and characterization of three new components of the mSin3A corepressor complex. Mol. Cell. Biol. 2003;23:3456–3467. doi: 10.1128/MCB.23.10.3456-3467.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, S.C., von Hippel, P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Hassig, C.A., Fleischer, T.C., Billin, A.N., Schreiber, S.L., Ayer, D.E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- Hurlin, P.J., Queva, C., Eisenman, R.N. Mnt: A novel Max-interacting protein and Myc antagonist. Curr. Top. Microbiol. Immunol. 1997;224:115–121. doi: 10.1007/978-3-642-60801-8_11. [DOI] [PubMed] [Google Scholar]
- Kim, H., Lee, J.E., Cho, E.J., Liu, J.O., Youn, H.D. Menin, a tumor suppressor, represses JunD-mediated transcriptional activity by association with an mSin3A-histone deacetylase complex. Cancer Res. 2003;63:6135–6139. [PubMed] [Google Scholar]
- Knoepfler, P.S., Eisenman, R.N. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- Koipally, J., Renold, A., Kim, J., Georgopoulos, K. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 1999;18:3090–3100. doi: 10.1093/emboj/18.11.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laherty, C.D., Yang, W.M., Sun, J.M., Davie, J.R., Seto, E., Eisenman, R.N. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- Laherty, C.D., Billin, A.N., Lavinsky, R.M., Yochum, G.S., Bush, A.C., Sun, J.M., Mullen, T.M., Davie, J.R., Rose, D.W., Glass, C.K., et al. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol. Cell. 1998;2:33–42. doi: 10.1016/s1097-2765(00)80111-2. [DOI] [PubMed] [Google Scholar]
- McArthur, G.A., Laherty, C.D., Queva, C., Hurlin, P.J., Loo, L., James, L., Grandori, C., Gallant, P., Shiio, Y., Hokanson, W.C., et al. The Mad protein family links transcriptional repression to cell differentiation. Cold Spring Harb. Symp. Quant. Biol. 1998;63:423–433. doi: 10.1101/sqb.1998.63.423. [DOI] [PubMed] [Google Scholar]
- Plevin, M.J., Mills, M.M., Ikura, M. The LxxLL motif: A multifunctional binding sequence in transcriptional regulation. Trends Biochem. Sci. 2005;30:66–69. doi: 10.1016/j.tibs.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Ptashne, M., Gann, A. Genes and signals. 1st ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2002. p. 192. [Google Scholar]
- Schreiber-Agus, N., DePinho, R.A. Repression by the Mad(Mxi1)–Sin3 complex. Bioessays. 1998;20:808–818. doi: 10.1002/(SICI)1521-1878(199810)20:10<808::AID-BIES6>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Silverstein, R.A., Ekwall, K. Sin3: A flexible regulator of global gene expression and genome stability. Curr. Genet. 2005;47:1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- Spronk, C.A., Tessari, M., Kaan, A.M., Jansen, J.F., Vermeulen, M., Stunnenberg, H.G., Vuister, G.W. The Mad1–Sin3B interaction involves a novel helical fold. Nat. Struct. Biol. 2000;7:1100–1104. doi: 10.1038/81944. [DOI] [PubMed] [Google Scholar]
- Stols, L., Gu, M., Dieckman, L., Raffen, R., Collart, F.R., Donnelly, M.I. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- Swanson, K.A., Knoepfler, P.S., Huang, K., Kang, R.S., Cowley, S.M., Laherty, C.D., Eisenman, R.N., Radhakrishnan, I. HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations. Nat. Struct. Mol. Biol. 2004;11:738–746. doi: 10.1038/nsmb798. [DOI] [PubMed] [Google Scholar]
- van Ingen, H., Lasonder, E., Jansen, J.F., Kaan, A.M., Spronk, C.A., Stunnenberg, H.G., Vuister, G.W. Extension of the binding motif of the Sin3 interacting domain of the Mad family proteins. Biochemistry. 2004;43:46–54. doi: 10.1021/bi0355645. [DOI] [PubMed] [Google Scholar]
- van Ingen, H., Baltussen, M.A., Aelen, J., Vuister, G.W. Role of structural and dynamical plasticity in Sin3: The free PAH2 domain is a folded module in mSin3B. J. Mol. Biol. 2006;358:485–497. doi: 10.1016/j.jmb.2006.01.100. [DOI] [PubMed] [Google Scholar]
- Wotton, D., Knoepfler, P.S., Laherty, C.D., Eisenman, R.N., Massague, J. The Smad transcriptional corepressor TGIF recruits mSin3. Cell Growth Differ. 2001;12:457–463. [PubMed] [Google Scholar]
- Yang, Q., Kong, Y., Rothermel, B., Garry, D.J., Bassel-Duby, R., Williams, R.S. The winged-helix/forkhead protein myocyte nuclear factor beta (MNF-β) forms a co-repressor complex with mammalian sin3B. Biochem. J. 2000;345:335–343. [PMC free article] [PubMed] [Google Scholar]
- Yochum, G.S., Ayer, D.E. Pf1, a novel PHD zinc finger protein that links the TLE corepressor to the mSin3A-histone deacetylase complex. Mol. Cell. Biol. 2001;21:4110–4118. doi: 10.1128/MCB.21.13.4110-4118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Sun, Z.W., Iratni, R., Erdjument-Bromage, H., Tempst, P., Hampsey, M., Reinberg, D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
- Zhang, J.S., Moncrieffe, M.C., Kaczynski, J., Ellenrieder, V., Prendergast, F.G., Urrutia, R. A conserved α-helical motif mediates the interaction of Sp1-like transcriptional repressors with the corepressor mSin3A. Mol. Cell. Biol. 2001;21:5041–5049. doi: 10.1128/MCB.21.15.5041-5049.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Zhang, Z., Demeler, B., Radhakrishnan, I. Coupled unfolding and dimerization by the PAH2 domain of the mammalian Sin3A corepressor. J. Mol. Biol. 2006;360:7–14. doi: 10.1016/j.jmb.2006.04.069. [DOI] [PubMed] [Google Scholar]