Abstract
Prions are proteins that can switch to self-perpetuating, infectious conformations. The abilities of prions to replicate, form structurally distinct strains, and to establish and overcome transmission barriers between species are poorly understood. We exploit surface-bound peptides to overcome complexities of investigating such problems in solution. For the yeast prion Sup35, we find that the switch to the prion state is controlled with exquisite specificity by small elements of primary sequence. Strikingly, these same sequence elements govern the formation of distinct self-perpetuating conformations (prion strains) and determine species-specific seeding activities. A Sup35 chimera that traverses the transmission barrier between two yeast species possesses the critical sequence elements from both. Using this chimera, we show that the influence of environment and mutations on the formation of species-specific strains is driven by selective recognition of one or the other sequence element. Thus, critical aspects of prion conversion are enciphered by subtle differences between small, highly-specific recognition elements.
The ability of proteins to form β-sheet rich amyloids is associated not only with disease1–3, but also with diverse normal biological functions including cell-adhesion4, skin pigmentation5, adaptation to environmental stresses6–8, and perhaps even long-term neuronal memory9. Prions are an unusual class of amyloid-forming proteins, whose conformations are self-templating (self-seeding) and thereby infectious. The conformationally-converted prion state can be transmitted from cell to cell within or, in some cases, between organisms. Prions, too, can be either deadly or beneficial6–8,10.
The first identified prion protein was PrP, whose conversion to the prion conformer (PrPSc) is associated with several fatal neurodegenerative diseases10. More recently identified prions in yeast and other fungi are unrelated to PrP or to one another11–14. Some of these have may beneficial effects6–8. The most well-studied is Sup3515–18, a translation-termination factor whose conversion to the prion state reduces its activity. This increases the read-through of stop codons, revealing hidden genetic variation and creating complex new phenotypes in a single step6–8. The ability of Sup35 to exist stably and heritably in either the prion or non-prion states, and to switch between them, constitutes a mechanism of epigenetic inheritance that is highly conserved across diverse fungal species19–24.
It is of great interest to determine how a protein functions as a genetic element. But investigations of Sup35 also provide insight into the conformational conversion of other amyloidogenic proteins. Sup35 was the first prion shown to be a self-templating amyloid25–27. Moreover, its assembly into prion (amyloid) conformers was discovered to involve an unusual conformationally-labile oligomeric intermediate28. It now appears that many other amyloid-forming proteins, including those involved in several human diseases, assemble via a similar intermediate29–35, and this oligomeric form may be a key factor in their pathogenicity36–43. Sup35 prions also exhibit two of the most baffling aspects of prion biology that were initially identified for mammalian PrP. First, both Sup35 and PrP can adopt not just one prion conformation, but a suite of related yet structurally distinct conformations (known as strains or variants)26,27,44–52. Each conformation self-perpetuates and confers a distinct biological phenotype. Second, the transmission of the prion state between proteins of different species is limited by a species barrier that can occasionally be traversed19–24,53–58. In both yeast and mammals the ability to establish and overcome species barriers is, in some unknown way, related to the ability of prions to form distinct strains10,15,53,56,58–65.
Sup35’s C-terminal domain encodes its translation-termination function15–17. Its ability to exist in either a prion or a non-prion state is controlled by two other domains15–17. The middle region (M) has a strong solubilizing activity and is very rich in charged residues. The amino-terminus (N) is extremely amyloidogenic and of unusually low sequence complexity, composed primarily of glutamine, asparagine, glycine and tyrosine residues (Supplementary Fig. S1). It is the interplay between these two domains (NM) that allows Sup35 to exist stably in either the prion (amyloid) state or the functional and soluble non-prion state.
In its non-prion state NM is compact, but molten, rapidly fluctuating through diverse conformations66. The structure of NM in its prion state is heavily debated. Several lines of evidence suggest that two discrete regions of the N domain are in self-contact within NM fibres67; the region between them is sequestered from intermolecular contacts, while elements proximal and distal to the contacts are not part of the amyloid core. Cross-linking NM molecules at one of the intermolecular contacts, but not elsewhere, accelerates nucleation67. However, other lines of evidence suggest that most residues of the N domain are in intermolecular contact, stacking in-register upon themselves68–72. Genetic evidence is also in conflict. Single substitution mutations in certain regions of N can have profound effects on many aspects of prion biology: they can inhibit replication73,74, bias prion conversion towards the production of distinct strains61,74, and increase or decrease the ability of prions to cross species barriers61. These studies would suggest that precise features of amino-acid sequence play critical roles in Sup35 prion biology. Remarkably, however, scrambling the sequence of N does not prevent prion formation71,72. This argues that NM prion formation is mainly dependent upon its amino-acid composition and largely independent of primary sequence71,72.
To shed light on these paradoxes, and to bring new technology to bear on the baffling relationship between prion strains and species barriers, we have employed surface-bound arrays of prion peptides. We find that small elements of the Saccharomyces cerevisiae Sup35 sequence govern prion recognition with extraordinary specificity. We exploited the properties of such sequences to identify critical prion recognition elements de novo in a distantly-related yeast species. Remarkably, the recognition elements identified using peptide arrays govern nucleation, the formation of distinct prion strains and the capacity of prions to cross species-specific transmission barriers.
Identification of high-specificity recognition elements using peptide arrays
We sought to determine how natively unfolded non-prion conformers achieve self-recognition. Is it encoded by specific sequence elements? Or is it a distributed property of the N domain’s unusual low-complexity sequence? To investigate this question we sought to interrogate the affinity of full-length proteins for diverse short elements of the NM sequence. However, short peptides from aggregation-prone proteins are often poorly soluble, precluding assessment of such interactions in solution.
To overcome this problem, we arrayed a library of overlapping denatured 20mer peptides derived from the sequence of the prion domain of S. cerevisiae Sup35 (ScNM) on glass slides. An extensive library of 136 overlapping peptides from ScNM was synthesized. Each peptide carried 20 residues of prion sequence at its C-terminus, preceded by a 14-carbon PEG spacer and an N-terminal, double lysine tag for covalent immobilization. The peptides, which should occupy ~2–3 nm in a random coil conformation under these conditions, were printed with an average spacing of ~10–12 nm to restrict self-interactions. Each peptide sequence was shifted from that of the preceding peptide by one to six residues. In the final version of the arrays smaller intervals were employed to provide higher resolution in critical regions that were identified in preliminary experiments. (The density of coverage is indicated by the spacing of the bars in the figures, with the x-axis referencing the centre of the peptide.) The ability of the immobilized peptides to interact with soluble, full-length NM was examined by incubating fluorescently labelled protein with the peptide arrays. The label was introduced via a cysteine substitution mutation (alanine to cysteine at residue 230, A230C) located in a region that lies far from the residues involved in prion formation67,73. The rest of the protein was identical to wild-type with no substitutions or labels.
Incubating full-length ScNM with the peptide arrays for two hours led to a strong accumulation of label over only a very small set of peptides: those encompassing amino acids (aa) 9–28, 10–29, 11–30, 12–31 and 20–39 (Fig. 1a, b and Supplementary Table S2). These interactions were highly reproducible (Fig 1a). Strikingly, these sequences lie within the region previously found by mutational analysis73 and crosslinking67 to strongly influence ScNM prion assembly. It also overlaps with one of two regions previously identified as being in self-contact within in mature fibres67.
Figure 1.
Identification of recognition sequences within ScNM using peptide arrays. a, Image of a triplicate array of sequential overlapping 20mer ScNM peptides after incubation with labelled full-length ScNM. b–c, Quantification of the fluorescence of labelled full-length ScNM bound to a similar peptide array after (b) two hours (1 μM, 5% Alexa Fluor 555) and (c) two and a half days (5 μM, 75% Alexa Fluor 555). The relative fluorescence intensity (RFU) for each 20mer peptide, reported as median + standard deviation, is displayed at its central residue on the x-axis. d–f, Quantification of the fluorescence of labelled full-length ScNM (1 μM, 5% Alexa Fluor 555) bound to ScNM peptides after (d) two hours, (e) one day and (f) one day after preincubation with unlabelled full-length ScNM (1 μM) for five days. g, Transmission electron micrograph of full-length ScNM fibres that assembled on a peptide array after two and a half days. The scale bar is 50 nm.
To determine if other peptide regions could interact with ScNM, albeit with lower efficiency, we took advantage of the fact that the spontaneous assembly of the full-length protein, even at five fold higher concentrations, is very slow in quiescent reactions28. Indeed, most of the protein remains soluble after two and a half days28. At this concentration and with a higher fraction of the protein carrying the fluorescent probe (75% vs. 5% of protein), label could be detected at a second set of peptides, spanning residues ~90–120 after one day (data not shown). This signal was very robust after two and a half days (Fig. 1c and Supplementary Table S2). This region corresponded to the second previously identified site of intermolecular contact within mature ScNM fibres (~85–105)67. The reactivity of the surface-bound peptides indicates that these regions are not only sites of intermolecular contact in mature fibres67, but also represent highly-specific self-recognition elements within soluble molten, non-prion conformers.
Recognition elements nucleate NM amyloid fibres
What is the nature of the protein bound to the peptide arrays? The fluorescence detected at specific peptide spots after two hours (Fig. 1d) continued to increase after even a full day of incubation (Fig. 1e). In a further attempt to saturate peptide reactivity on the arrays, the slides were pre-incubated with unlabelled ScNM for five days. However, when subsequently incubated with labelled protein, these spots continued to accumulate label as above (Fig. 1f).
These results suggested that the growing signals at specific peptide spots might not simply be due to slow recognition of rare molten soluble conformers. Rather, they might represent sites of prion assembly. Indeed, the fluorescent signal of the bound ScNM was resistant to extensive washing with 2% SDS, a characteristic of the prion amyloids formed by NM28,75. To test the nature of the material accumulating at these specific spots more directly, the protein was scraped from the slide and imaged by transmission electron microscopy (Fig. 1g). Abundant fibres were observed, and they were of the characteristic diameter of NM prion fibres25.
The concentration of peptides in the arrayed spots is ~300-fold greater than the protein in bulk solution. Thus, it would seem reasonable to suppose that soluble proteins achieved critical nucleating contacts with peptides on the arrays more rapidly than they did with other full-length proteins in solution, which are presenting a diversity of sequences for interaction. However, the striking correspondence between the peptides at which fibres accumulated and the amino-acid segments that form intermolecular contacts in mature ScNM amyloids67 suggests an alternative explanation: the peptides simply captured ScNM amyloids that were assembling in solution during the incubation. To test this, labelled pre-assembled ScNM amyloid fibres were sonicated into small fragments and incubated with the arrays. In this case the fluorescence that accumulated at the peptide spots was broadly distributed across the array and similar to background (data not shown). We conclude that the fibres associated with specific peptide spots have assembled there in situ from soluble NM. It was not possible for us to determine whether the interacting soluble species is monomeric or oligomeric. Regardless, our data clearly indicate that with an extraordinary degree of specificity small elements of the Sup35 prion sequence are able to recognize molten conformers of full-length NM and convert them via this recognition to a self-templating state.
C. albicans NM interacts with short peptides in a similar manner
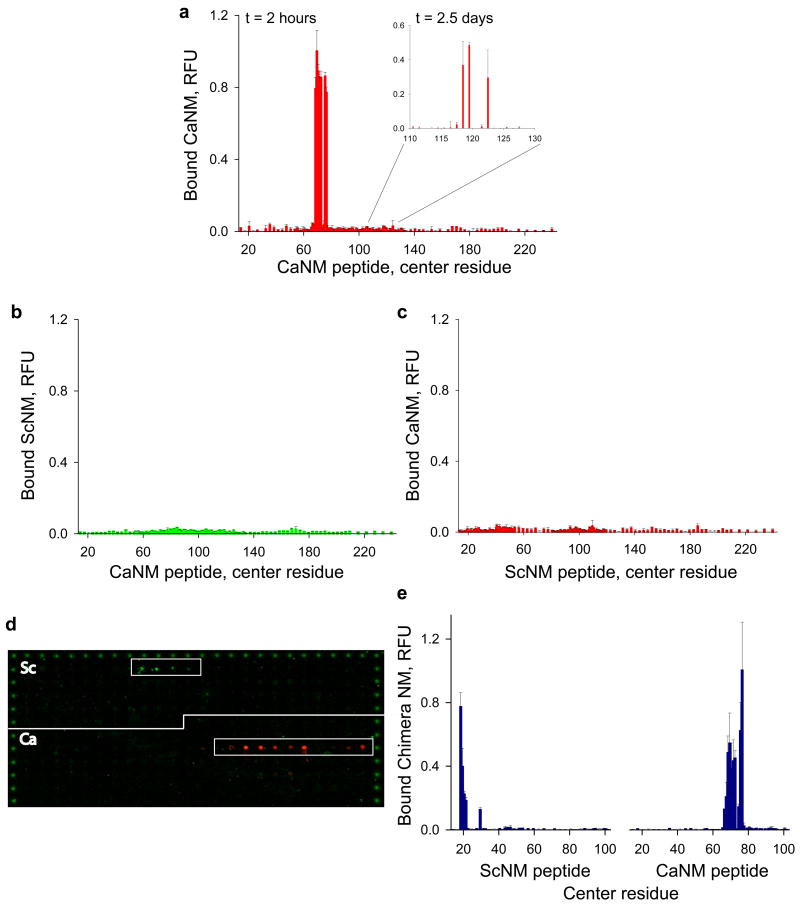
Next, we asked if another prion protein might have specific, highly-localized recognition elements and if our method could be used to identify them. We employed the homologous prion-determining region of Sup35 from C. albicans, a species separated from S. cerevisiae by >800 million years of evolution76. The sequence of C. albicans NM (CaNM) shares no significant tracts of alignable sequence with S. cerevisiae NM24, but is of similar low complexity and amino-acid composition (Supplementary Fig. S1). CaNM was fluorescently labelled as for ScNM and incubated with a library of 128 overlapping CaNM peptides. After two hours CaNM reacted in a highly-specific manner with a small cluster of peptides: aa 59–78, 60–79, 61–80, 62–81, 63–82, 66–85 and 67–86 (Fig. 2a and Supplementary Table S3). After longer incubations (one day) at five-fold higher CaNM concentrations, and with a higher fraction of the protein carrying the fluorescent probe (75% vs. 5% of protein), label accumulated at a second set of peptides, spanning residues ~110–130 (data not shown). This signal was very robust after two and a half days (Fig. 2a, inset and Supplementary Table S3). As with ScNM, the signals obtained with CaNM were not saturable and continued to increase over several days of incubation (data not shown). Further, the signals were resistant to extensive washing with 2% SDS. Remarkably, for two distantly-related yeast species, short peptides from two distinct regions of the prion domain display a similar pattern of recognition by the full-length soluble proteins and a similar capacity to convert them, via this recognition, to a self-templating state.
Figure 2.
Analysis of CaNM recognition sequences and the species barrier between Sc/Ca NM. a, Quantification of the fluorescence of labelled full-length CaNM bound to overlapping 20mer CaNM peptides after two hours (1 μM, 5% Alexa Fluor 647) and two and a half days (inset, 5 μM, 75% Alexa Fluor 647). b, Quantification of the fluorescence of labelled full-length ScNM (1 μM, 5% Alexa Fluor 555) bound to overlapping 20mer CaNM peptides after two hours. c, Quantification of the fluorescence of labelled full-length CaNM (1 μM, 5% Alexa Fluor 647) bound to overlapping 20mer ScNM peptides after two hours. d, Image of a single peptide array that was simultaneously incubated with labelled full-length ScNM (green, Alexa Fluor 555) and CaNM (red, Alexa Fluor 647) for two hours (1 μM final concentration of each protein, 5% label). e, Quantification of the fluorescence of labelled full-length Sc/Ca NM chimera (1 μM NM, 5% Alexa Fluor 647) bound to both ScNM and CaNM peptides after two hours of incubation.
Reconstitution of the species barrier on peptide arrays
A strong species barrier exists between Sc and Ca Sup35 prions19,24. Prion fibres of each species can seed the polymerization of its own protein in vitro and can propagate their own prions in vivo, but each cross-seeds the protein of the other species very inefficiently19,24,59,61. The striking specificity by which specific peptides interact with and nucleate assembly of full-length ScNM and CaNM prompted us to ask if the species barrier could be explained by the recognition elements uncovered by the peptide arrays. Indeed, when labelled full-length ScNM was incubated with a peptide array displaying 128 CaNM peptides for two hours, no association was detected above background (Fig. 2b). The interaction of ScNM with peptides in its own recognition element (encompassing residues 9–39) after two hours was at least 30 fold greater than its interaction with any of the CaNM peptides. Conversely, incubating labelled full-length CaNM with an array displaying 136 ScNM peptides for two hours produced only background levels of fluorescence (Fig. 2c). The interaction of CaNM with its own peptides (encompassing residues 59–86) after two hours was 28 fold greater than for any of the ScNM peptides. The high degree of specificity that ScNM and CaNM displayed for their own peptides was maintained after longer incubation times (2.5 days, data not shown).
To test the strength of the species barrier observed on the peptide arrays more stringently, we mixed proteins of both species, each labelled with a different colour dye (ScNM, green and CaNM, red) and incubated them with arrays containing both peptides. The NM proteins of each species were highly selective for interaction with a small subset of peptides from their own sequences (Fig. 2d). No co-localization of labels was detected.
Next, we employed a promiscuous Sc/Ca NM chimera that has been shown previously to traverse the species barrier between S. cerevisiae and C. albicans19,61. This chimeric protein contains segments from both ScNM and CaNM (residues Sc 1–40, Ca 49–141 and Sc 124–253; Supplementary Fig. S1)19,61. Incubating the full-length NM chimera with an array displaying libraries of both ScNM and CaNM peptides revealed that it was able to interact with the prion recognition elements from both species in a highly-specific manner (Fig. 2e).
Role of recognition elements in the formation of prion strains with species-specific seeding activities
These results suggest that 1) the species barrier between ScNM and CaNM can be defined in terms of the differential affinities of soluble protein for specific short sequence elements that have the capacity to initiate a self-templating state, and 2) the ability to cross this species barrier is linked to the ability of soluble protein to interact promiscuously with sequence elements from both species. To test these hypotheses more rigorously, we exploited the ability of the Sc/Ca NM chimera to assemble into distinct strains that have species-specific seeding activities19,59,61. Protein fibres assembled from the chimeric protein at 15°C seed the amyloid formation of ScNM but not CaNM61. Fibres assembled at 37°C have the opposite specificity61. For ease of manipulation we confirmed that the species-specific strain properties obtained at 15°C were also obtained at 4°C at the protein concentrations employed for our arrays (data not shown).
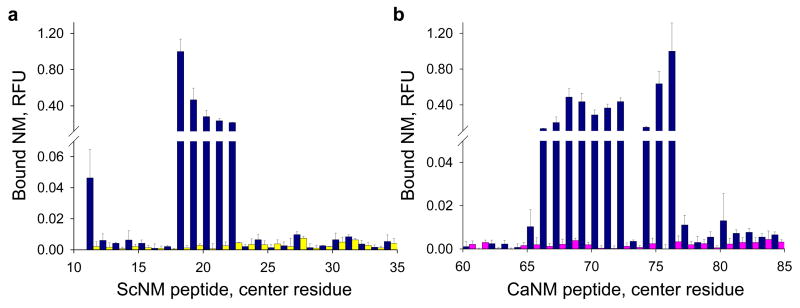
As described above, when the chimeric protein was incubated with the peptide arrays at 25°C, it interacted with peptides from both species (Fig. 2e). However, at 37°C it interacted selectively with CaNM peptides (Fig. 3a). At 4°C the chimera interacted selectively with ScNM peptides (Fig. 3b). Thus, the ability of the chimeric protein to assemble into distinct species-specific strains at different temperatures is enciphered by the same small sequence elements that nucleate amyloid assembly.
Figure 3.
Analysis of the conformational preference of the Sc/Ca NM chimera. a–d, Quantification of the relative binding of various labelled full-length NM chimeric proteins to overlapping 20mer ScNM and CaNM peptides: (a) Sc/Ca chimera at 37°C, (b) Sc/Ca chimera at 4°C, (c) S17R Sc/Ca chimera at 25°C and (d) G70, 71, 80, 81A Sc/Ca chimera at 25°C. The peptide arrays were incubated with each prion domain (1 μM NM, 5% Alexa Fluor 647) for two hours.
Next, we tested the hypothesis that the effects of mutations on the formation of species-specific strains could also be explained by these same prion recognition elements. As previously reported, changing a serine at residue 17 to an arginine (S17R) in the Sc/Ca chimera favours assembly of a prion strain that selectively seeds CaNM61. Conversely, changing four glycine residues at positions 70, 71, 80 and 81 to alanine (4G/A) favours assembly of a prion strain that selectively seeds ScNM61. When chimeric protein with the S17R mutation was incubated with the peptide arrays at 25°C, binding to all of the ScNM peptides was reduced to background but binding to CaNM peptides was unaffected (Fig. 3c). When the 4G/A mutant was incubated with the arrays, binding to all but one of the CaNM peptides was greatly reduced but binding to ScNM peptides was similar to the original chimera (Fig. 3d).
Mechanism of mutational disruption of Sup35 recognition elements
How do the mutations in the Sc/Ca chimera alter prion recognition and strain formation? They might do so by biasing the conformations sampled by the molten full-length proteins such that particular recognition elements are masked. Alternatively, the mutations may directly interfere with interactions between the full-length proteins and their cognate recognition elements. To investigate this question we employed the original chimeric protein and tested its ability to interact with arrays containing mutant peptides. The labelled chimeric protein bound robustly to the wild-type ScNM and CaNM peptides (dark blue bars, Fig. 4a and b), but exhibited no interaction above background with any ScNM peptides containing the S17R mutation (yellow bars, Fig. 4a) or any CaNM peptides containing the 4G/A mutations (pink bars, Fig. 4b). Since the original chimera reacted with wild-type peptides on the same array, it must have displayed its own recognition element. Its inability to interact with the mutant peptides, therefore, indicates that the mutations directly disrupt the recognition function of the sequence elements rather than solely altering the conformations of the soluble protein. Thus, these mutations, which bias prions toward the formation of distinct strains and alter cross-species prion transmission, do so by directly interfering with recognition of the prion specificity elements.
Figure 4.
Analysis of the mutational disruption of the ScNM and CaNM recognition elements. a, Quantification of the relative affinity of the full-length Sc/Ca NM chimera for wild-type ScNM peptides (dark blue bars) and ScNM peptides containing the S17R mutation (yellow bars). b, Quantification of the relative affinity of the full-length Sc/Ca NM chimera for wild-type CaNM peptides (dark blue bars) and CaNM peptides containing the G70, 71, 80, 81A mutations (pink bars). The peptide arrays were incubated with the NM chimera (1 μM NM, 5% Alexa Fluor 647) for two hours.
Discussion
Using arrays of surface-bound peptides from the prion domains of S. cerevisiae and C. albicans Sup35 proteins, we find that small elements of sequence govern self-recognition with exquisite specificity. These recognition elements are alone sufficient to drive the conversion of full-length proteins in the non-prion conformation – molten, rapidly fluctuating, natively unfolded conformers – into self-templating amyloid conformations. Moreover, it is these same sequence elements that govern the formation of the distinct prion strains and determine transmission barriers between species.
The highly-localized and species-specific nature of the interactions detected with peptide arrays was unexpected for several reasons. For example, when the sequence of S. cerevisiae’s Sup35 prion domain is scrambled, the protein still forms prions71,72. This had suggested that prion formation was a distributed property, due to the unusual amino-acid composition of the N domain, rather than a localized property of specific sequences within it. Indeed, scrambling of the prion domain sequence has occurred in nature for different yeast species. Although the S. cerevisiae and C. albicans N domains are both of low complexity (half of their residues are glutamines or asparagines and most others are tyrosines or glycines; Supplementary Fig. S1), their amino-acid sequences cannot be aligned24. Yet CaNM forms prions with a similar efficiency as ScNM19,24. This, too, might suggest that prion formation is a distributed property governed primarily by amino-acid composition.
We have found, however, that full-length ScNM and CaNM interact with only a small number of peptides derived from their own sequences and do so in a highly-localized and specific manner. But how can our data be reconciled with the ability of scrambled sequences to produce bona fide heritable prion elements? One explanation is that the scrambled NM sequences may form prions at very low efficiency; they were expressed at very high levels and their rates of prion conversion relative to the wild-type were not reported71. Alternatively, it may be that scrambling N domain sequences frequently results in the formation of new recognition elements. Given that the prion state produces many changes in phenotype (some detrimental and some beneficial)6–8, the efficiency of prion conversion may have been conserved for ScNM and CaNM through selective pressures despite sequence scrambling.
The peptides in the recognition elements we identified also have the unique capacity to drive molten non-prion conformers into a self-templating state. It has recently been reported that small amyloidogenic peptides from the N domain of ScNM (residues 7–12 and 7–13, which partially overlap with one of our recognition elements) can form microcrystals that yield high-resolution structures77. Our identification of critical nucleating peptides, therefore, holds promise for future high-resolution analysis of the bona fide templating structures of [PSI+], and potentially the comparative analysis of such structures from organisms separated by 800 million years of evolution.
Remarkably, the non-prion conformers of both ScNM and CaNM interacted initially and most strongly with a small cluster of their own peptides towards the N-terminal side of the N domain (Fig. 1b and 2a). Additionally, both proteins could interact with a second discrete cluster of their own peptides towards the C-terminal end of the N domain, but only after much longer incubations and at higher protein concentrations (Fig 1c and 2a, inset). This finding suggests that the nature of prion nucleation, conformational conversion and templating for Sup35 may have been conserved during evolution. If true, this would provide strong evidence for our previous hypothesis that the phenotypic variation created when Sup35 switches to the prion state is of considerable biological importance6,7.
It is striking that the specific recognition sites identified on the arrays correspond to regions of intermolecular association identified in ScNM fibres by other methods67. Although our results cannot distinguish between proposed structural models for ScNM prion fibres67–70, the data seem most consistent with one of them: that two distinct regions in the N domain are in self-contact, with the intervening region sequestered from intermolecular contacts, and regions distal and proximal to the contacts being outside the amyloid core67. They are also consistent with cross-linking data suggesting that one of the two contact sites plays a dominant role in nucleation67.
Our data also suggest a new framework for conceptualizing the formation of species barriers and prion strains. Species barriers are known to depend, in part, upon the primary sequence of prions19–24,53–58. We find that they are determined by highly localized elements of amino-acid sequence. Moreover, these are the very same elements that control nucleation. Species barriers are also known to be influenced by prion strains10,15,53,56,58–65, but the molecular connection has been elusive. For the promiscuous Sc/Ca NM chimera the remarkable concordance between the way that specific mutations and environmental conditions determine the particular peptides that can nucleate soluble chimeric protein on the arrays (Fig. 3) and way they determine the species-specific propagation of different chimeric strains61 suggests the following relationship: within the same region of the same protein, small elements of primary sequence can initiate distinct intermolecular recognition events that drive the formation of different amyloid structures. These same sequence elements are then involved in templating those structures to soluble protein for propagation of the prion strain.
From a methodological perspective, it is empowering that peptide arrays, which are typically used to study the specificities of folded proteins78, can also be used to identify sequences that drive structural transitions in natively unfolded proteins. This method has the potential to illuminate many difficult problems in human disease biology. For example, genetic analysis of prion strain variation and species barriers has been far less tractable in mammals than in yeast. Peptide arrays may prove equally applicable to both, allowing testable hypotheses to be rapidly generated. Peptide arrays might also be employed in high-throughput drug screens for compounds that prevent protein assembly, promote it, or redirect it to alternative forms. Finally, the fact that highly-ordered amyloid fibres can be specifically nucleated and assembled at patterned sites on solid surfaces, and that this assembly process can be tightly controlled by simple changes in environmental conditions (e.g., temperature), may have important implications for fabricating materials and devices with complex, nanostructured features79,80.
Perhaps the most important implication of our work is that the very nature of protein folding for Sup35 prions is profoundly different from that of proteins with globular domains. The latter is typically governed by a large number of intramolecular interactions that collectively and cooperatively drive folding of the entire domain. In contrast, we find that the folding of two distantly-related yeast prion proteins into amyloid fibres of defined tertiary structure is controlled by intermolecular interactions between very small elements of primary sequence. Peptide arrays offer an opportunity to determine whether other amyloidogenic proteins fold by a similar mechanism.
Methods
Mutagenesis, protein purification and cysteine labeling
Single cysteine mutations were introduced into NM using QuikChange mutagenesis (Stratagene). ScNM and CaNM contained a C-terminal 7x His tag, as did the Sc/Ca NM chimera constructs except for S17R, which contained a 6x His tag. All NM proteins were purified as described previously25 except that the proteins were eluted from a Ni-NTA column using low pH instead of imidazole. The NM cysteine mutants at or near the C-terminus (Sc A230C, Ca S227C and all chimeras at the extreme C-terminus) were labelled overnight at room temperature with maleimide-functionalized Alexa Fluor 555 or 647 (Invitrogen) using a 5:1 to 10:1 molar ratio of label:NM, and the free label was removed using a Ni-NTA column.
Peptide array synthesis, hybridization and quantification
Peptides were synthesized on modified cellulose membranes using SPOT technology81 (JPT Peptide Technologies GmbH). Each peptide contained a double alanine tag at its N-terminus, a 20-residue segment derived from the Sup35 sequences, a hydrophilic linker (1-amino-4,7,10-trioxa-13-tridecanamine succinimic acid82) and a double lysine tag at its C-terminus. The peptides were cleaved off the membranes, freeze-dried and resuspended in buffer (40% DMSO, 5% glycerol, 55% PBS, pH 9) for printing. The peptides were then printed onto hydrogel glass slides (Nexterion Slide H, Schott) functionalized with reactive N-hydroxysuccinimide (NHS) ester moieties. Each peptide spot (250 μm in diameter) was printed with 1.5 nl of peptide solution at a concentration of approximately 2.5 μM using non-contact printing (JPT Peptide Technologies GmbH). Unreacted peptides were removed from the hydrogel slides. Slides were dried and blocked with 3% BSA in PBST for 1 h. The NM proteins were denatured in 6 M guanidine hydrochloride at 100°C for approximately 20 min, and then diluted 125 fold in PBST containing 3% BSA to a final concentration of 1–5 μM NM and a label-per-protein molar ratio of 5–75%. Each peptide array was incubated individually with 2–3 ml of diluted NM in an Altas hybridization chamber (BD Biosciences) for the indicated times without mixing. The peptide arrays were then washed 5 times with 50 ml of 2% SDS for 30 min, 5 times with 50 ml of water, 3 times with 50 ml of methanol and then spun dry. The methanol washes were not essential but helped prevent uneven drying of the slides. The arrays were then imaged using a Genepix 4000A scanner and the median fluorescence values of two or three replicates for the peptide spots were quantified using Genepix Pro 6.0 software (Molecular Devices).
Supplementary Material
Acknowledgments
We thank Jonathan Weissman for providing the CaNM and Sc/Ca NM chimera plasmids, Mike Schutkowski for assistance in designing and preparing the peptide arrays, Nicki Watson for performing the TEM imaging and members of the Lindquist lab for helpful discussions. This research was supported by an American Cancer Society Postdoctoral Fellowship (P.M.T.), and grants from the DuPont-MIT Alliance and the NIH (NIGMS 25874).
References
- 1.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Sacchettini JC, Kelly JW. Therapeutic strategies for human amyloid diseases. Nat Rev Drug Discov. 2002;1:267–275. doi: 10.1038/nrd769. [DOI] [PubMed] [Google Scholar]
- 3.Selkoe DJ. Folding proteins in fatal ways. Nature. 2003;426:900–904. doi: 10.1038/nature02264. [DOI] [PubMed] [Google Scholar]
- 4.Chapman MR, et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fowler DM, et al. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:100–107. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 7.True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–187. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- 8.Eaglestone SS, Cox BS, Tuite MF. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. Embo J. 1999;18:1974–1981. doi: 10.1093/emboj/18.7.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Si K, Lindquist S, Kandel ER. A neuronal isoform of the Aplysia CPEB has prion-like properties. Cell. 2003;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- 10.Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wickner RB, Masison DC. Evidence for two prions in yeast: [URE3] and [PSI] Curr Top Microbiol Immunol. 1996;207:147–160. doi: 10.1007/978-3-642-60983-1_10. [DOI] [PubMed] [Google Scholar]
- 12.Wickner RB. [Ure3] as an altered Ure2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 13.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 14.Coustou V, Deleu C, Saupe S, Begueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci U S A. 1997;94:9773–9778. doi: 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chien P, Weissman JS, DePace AH. Emerging principles of conformation-based prion inheritance. Annu Rev Biochem. 2004;73:617–656. doi: 10.1146/annurev.biochem.72.121801.161837. [DOI] [PubMed] [Google Scholar]
- 16.Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- 17.Tuite MF, Cox BS. Propagation of yeast prions. Nat Rev Mol Cell Biol. 2003;4:878–890. doi: 10.1038/nrm1247. [DOI] [PubMed] [Google Scholar]
- 18.Uptain SM, Lindquist S. Prions as protein-based genetic elements. Annu Rev Microbiol. 2002;56:703–741. doi: 10.1146/annurev.micro.56.013002.100603. [DOI] [PubMed] [Google Scholar]
- 19.Chien P, Weissman JS. Conformational diversity in a yeast prion dictates its seeding specificity. Nature. 2001;410:223–227. doi: 10.1038/35065632. [DOI] [PubMed] [Google Scholar]
- 20.Chernoff YO, et al. Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol Microbiol. 2000;35:865–876. doi: 10.1046/j.1365-2958.2000.01761.x. [DOI] [PubMed] [Google Scholar]
- 21.Kushnirov VV, Kochneva-Pervukhova NV, Chechenova MB, Frolova NS, Ter-Avanesyan MD. Prion properties of the Sup35 protein of yeast Pichia methanolica. Embo J. 2000;19:324–331. doi: 10.1093/emboj/19.3.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakayashiki T, Ebihara K, Bannai H, Nakamura Y. Yeast [PSI+] “prions” that are crosstransmissible and susceptible beyond a species barrier through a quasi-prion state. Mol Cell. 2001;7:1121–1130. doi: 10.1016/s1097-2765(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 23.Resende C, et al. The Candida albicans Sup35p protein (CaSup35p): function, prion-like behaviour and an associated polyglutamine length polymorphism. Microbiology. 2002;148:1049–1060. doi: 10.1099/00221287-148-4-1049. [DOI] [PubMed] [Google Scholar]
- 24.Santoso A, Chien P, Osherovich LZ, Weissman JS. Molecular basis of a yeast prion species barrier. Cell. 2000;100:277–288. doi: 10.1016/s0092-8674(00)81565-2. [DOI] [PubMed] [Google Scholar]
- 25.Glover JR, et al. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 26.King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 28.Serio TR, et al. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 29.Nettleton EJ, et al. Characterization of the oligomeric states of insulin in self-assembly and amyloid fibril formation by mass spectrometry. Biophys J. 2000;79:1053–1065. doi: 10.1016/S0006-3495(00)76359-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldberg MS, Lansbury PT., Jr Is there a cause-and-effect relationship between alpha-synuclein fibrillization and Parkinson’s disease? Nat Cell Biol. 2000;2:115–119. doi: 10.1038/35017124. [DOI] [PubMed] [Google Scholar]
- 31.Eakin CM, Attenello FJ, Morgan CJ, Miranker AD. Oligomeric assembly of native-like precursors precedes amyloid formation by beta-2 microglobulin. Biochemistry. 2004;43:7808–7815. doi: 10.1021/bi049792q. [DOI] [PubMed] [Google Scholar]
- 32.Conway KA, et al. Acceleration of oligomerization, not fibrillization, is a shared property of both alpha-synuclein mutations linked to early-onset Parkinson’s disease: implications for pathogenesis and therapy. Proc Natl Acad Sci U S A. 2000;97:571–576. doi: 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kayed R, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 34.Shorter J, Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell. 2006;23:425–438. doi: 10.1016/j.molcel.2006.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bader R, Bamford R, Zurdo J, Luisi BF, Dobson CM. Probing the mechanism of amyloidogenesis through a tandem repeat of the PI3-SH3 domain suggests a generic model for protein aggregation and fibril formation. J Mol Biol. 2006;356:189–208. doi: 10.1016/j.jmb.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 36.Silveira JR, et al. The most infectious prion protein particles. Nature. 2005;437:257–261. doi: 10.1038/nature03989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bucciantini M, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 38.Walsh DM, et al. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 39.Lesne S, et al. A specific amyloid-beta protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 40.Lambert MP, et al. Diffusible, nonfibrillar ligands derived from Abeta1–42 are potent central nervous system neurotoxins. Proc Natl Acad Sci U S A. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sousa MM, Cardoso I, Fernandes R, Guimaraes A, Saraiva MJ. Deposition of transthyretin in early stages of familial amyloidotic polyneuropathy: evidence for toxicity of nonfibrillar aggregates. Am J Pathol. 2001;159:1993–2000. doi: 10.1016/s0002-9440(10)63050-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cleary JP, et al. Natural oligomers of the amyloid-beta protein specifically disrupt cognitive function. Nat Neurosci. 2005;8:79–84. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- 43.Baglioni S, et al. Prefibrillar amyloid aggregates could be generic toxins in higher organisms. J Neurosci. 2006;26:8160–8167. doi: 10.1523/JNEUROSCI.4809-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pattison IH, Millson GC. Scrapie produced experimentally in goats with special reference to the clinical syndrome. J Comp Pathol. 1961;71:101–109. doi: 10.1016/s0368-1742(61)80013-1. [DOI] [PubMed] [Google Scholar]
- 45.Bruce ME, McConnell I, Fraser H, Dickinson AG. The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J Gen Virol. 1991;72:595–603. doi: 10.1099/0022-1317-72-3-595. [DOI] [PubMed] [Google Scholar]
- 46.Legname G, et al. Strain-specified characteristics of mouse synthetic prions. Proc Natl Acad Sci U S A. 2005;102:2168–2173. doi: 10.1073/pnas.0409079102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caughey B, Raymond GJ, Bessen RA. Strain-dependent differences in beta-sheet conformations of abnormal prion protein. J Biol Chem. 1998;273:32230–32235. doi: 10.1074/jbc.273.48.32230. [DOI] [PubMed] [Google Scholar]
- 48.Kocisko DA, et al. Cell-free formation of protease-resistant prion protein. Nature. 1994;370:471–474. doi: 10.1038/370471a0. [DOI] [PubMed] [Google Scholar]
- 49.Somerville RA, et al. Characterization of thermodynamic diversity between transmissible spongiform encephalopathy agent strains and its theoretical implications. J Biol Chem. 2002;277:11084–11089. doi: 10.1074/jbc.M111766200. [DOI] [PubMed] [Google Scholar]
- 50.Safar J, et al. Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 51.Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chernoff YO, Derkach IL, Inge-Vechtomov SG. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet. 1993;24:268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- 53.Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–550. doi: 10.1146/annurev.neuro.24.1.519. [DOI] [PubMed] [Google Scholar]
- 54.Scott M, et al. Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell. 1989;59:847–857. doi: 10.1016/0092-8674(89)90608-9. [DOI] [PubMed] [Google Scholar]
- 55.Prusiner SB, et al. Transgenic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 56.Collinge J, et al. Unaltered susceptibility to BSE in transgenic mice expressing human prion protein. Nature. 1995;378:779–783. doi: 10.1038/378779a0. [DOI] [PubMed] [Google Scholar]
- 57.Supattapone S, et al. Prion protein of 106 residues creates an artificial transmission barrier for prion replication in transgenic mice. Cell. 1999;96:869–878. doi: 10.1016/s0092-8674(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 58.Bruce M, et al. Transmission of bovine spongiform encephalopathy and scrapie to mice: strain variation and the species barrier. Philos Trans R Soc Lond B Biol Sci. 1994;343:405–411. doi: 10.1098/rstb.1994.0036. [DOI] [PubMed] [Google Scholar]
- 59.Tanaka M, Chien P, Yonekura K, Weissman JS. Mechanism of cross-species prion transmission: An infectious conformation compatible with two highly divergent yeast prion proteins. Cell. 2005;121:49–62. doi: 10.1016/j.cell.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 60.Hill AF, et al. The same prion strain causes vCJD and BSE. Nature. 1997;389:448–450. doi: 10.1038/38925. [DOI] [PubMed] [Google Scholar]
- 61.Chien P, DePace AH, Collins SR, Weissman JS. Generation of prion transmission barriers by mutational control of amyloid conformations. Nature. 2003;424:948–951. doi: 10.1038/nature01894. [DOI] [PubMed] [Google Scholar]
- 62.Collinge J, Sidle KC, Meads J, Ironside J, Hill AF. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature. 1996;383:685–690. doi: 10.1038/383685a0. [DOI] [PubMed] [Google Scholar]
- 63.Dickinson AG, Fraser H. Modification of the pathogenesis of scrapie in mice by treatment of the agent. Nature. 1969;222:892–893. doi: 10.1038/222892a0. [DOI] [PubMed] [Google Scholar]
- 64.Will RG, Ironside JW, Hornlimann B, Zeidler M. Creutzfeldt-Jakob disease. Lancet. 1996;347:65–66. [PubMed] [Google Scholar]
- 65.Peretz D, et al. A change in the conformation of prions accompanies the emergence of a new prion strain. Neuron. 2002;34:921–932. doi: 10.1016/s0896-6273(02)00726-2. [DOI] [PubMed] [Google Scholar]
- 66.Mukhopadhyay S, Krishnan R, Lemke EA, Lindquist S, Deniz AA. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc Natl Acad Sci U S A. 2007 doi: 10.1073/pnas.0611503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kajava AV, Baxa U, Wickner RB, Steven AC. A model for Ure2p prion filaments and other amyloids: the parallel superpleated beta-structure. Proc Natl Acad Sci U S A. 2004;101:7885–7890. doi: 10.1073/pnas.0402427101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc Natl Acad Sci U S A. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ross ED, Minton A, Wickner RB. Prion domains: sequences, structures and interactions. Nat Cell Biol. 2005;7:1039–1044. doi: 10.1038/ncb1105-1039. [DOI] [PubMed] [Google Scholar]
- 71.Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc Natl Acad Sci U S A. 2005;102:12825–12830. doi: 10.1073/pnas.0506136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ross ED, Baxa U, Wickner RB. Scrambled prion domains form prions and amyloid. Mol Cell Biol. 2004;24:7206–7213. doi: 10.1128/MCB.24.16.7206-7213.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.DePace AH, Santoso A, Hillner P, Weissman JS. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell. 1998;93:1241–1252. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- 74.King CY. Supporting the structural basis of prion strains: induction and identification of [PSI] variants. J Mol Biol. 2001;307:1247–1260. doi: 10.1006/jmbi.2001.4542. [DOI] [PubMed] [Google Scholar]
- 75.Serio TR, Cashikar AG, Moslehi JJ, Kowal AS, Lindquist SL. Yeast prion [PSI+] and its determinant, Sup35p. Methods Enzymol. 1999;309:649–673. doi: 10.1016/s0076-6879(99)09043-6. [DOI] [PubMed] [Google Scholar]
- 76.Heckman DS, et al. Molecular evidence for the early colonization of land by fungi and plants. Science. 2001;293:1129–1133. doi: 10.1126/science.1061457. [DOI] [PubMed] [Google Scholar]
- 77.Nelson R, et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eichler J. Synthetic peptide arrays and peptide combinatorial libraries for the exploration of protein-ligand interactions and the design of protein inhibitors. Comb Chem High Throughput Screen. 2005;8:135–143. doi: 10.2174/1386207053258497. [DOI] [PubMed] [Google Scholar]
- 79.Zhao XM, Xia YN, Whitesides GM. Soft lithographic methods for nano-fabrication. J Mater Chem. 1997;7:1069–1074. [Google Scholar]
- 80.Scheibel T, et al. Conducting nanowires built by controlled self-assembly of amyloid fibers and selective metal deposition. Proc Natl Acad Sci U S A. 2003;100:4527–4532. doi: 10.1073/pnas.0431081100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frank R. The SPOT-synthesis technique. Synthetic peptide arrays on membrane supports-principles and applications. J Immunol Methods. 2002;267:13–26. doi: 10.1016/s0022-1759(02)00137-0. [DOI] [PubMed] [Google Scholar]
- 82.Zhao ZG, Im JS, Lam KS, Lake DF. Site-specific modification of a single-chain antibody using a novel glyoxylyl-based labeling reagent. Bioconjugate Chem. 1999;10:424–430. doi: 10.1021/bc980120k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.