Summary
Common fragile sites are regions of human chromosomes prone to breakage. Fragile site FRA16D spans the WWOX/FOR tumor suppressor gene and has been linked to cancer-causing deletions and translocations. Using a genetic assay in yeast, we found that a short AT-rich region (Flex1) within FRA16D increases chromosome fragility whereas three other sequences within FRA16D do not. This is the first identification of a sequence element within a common fragile site that increases chromosome fragility. The fragility of Flex1 was exacerbated by the absence of Rad52 or presence of hydroxyurea. Flex1 contains a polymorphic AT repeat predicted to form a DNA structure, and two-dimensional gel analysis showed accumulation of stalled replication forks at the Flex1 sequence that was dependent on AT length. Our data suggest that the FRA16D Flex1 sequence causes increased chromosome breakage by forming secondary structures that stall replication fork progression.
Keywords: chromosome fragility, chromosome breakage, replication fork stalling, common fragile site, DNA structure, hairpin, cruciform
Introduction
Common fragile sites are chromosomal regions especially prone to forming gaps and breaks after treatment of cells with replication inhibitors such as aphidicolin, an inhibitor of polymerase α, δ and ε (Richards, 2001). These gaps and breaks are visible cytogenetically on metaphase chromosomes of treated cells. Common fragile sites can be induced in a large proportion of individuals and are therefore considered a normal component of chromosome structure. Over 80 common fragile sites have been identified after aphidicolin treatment, with the 20 most highly expressed sites accounting for over 80% of breaks (Glover et al., 2005). Genetic instability in vivo is a shared feature of common fragile sites. They are hot spots for sister chromatid exchanges (Glover and Stein, 1987), preferred sites for viral integration (Mishmar et al., 1998; Wang et al., 1992) and they can initiate adjacent gene amplification by a breakage-fusion-bridge mechanism (Coquelle et al., 1998). Most importantly, frequent deletions, loss of heterozygosity and translocations have been observed at common fragile site sequences in various tumor cells (Huebner and Croce, 2001). Some of these rearrangements may result in cancer, for example, about 25% of cases of multiple myeloma are caused by a t(14;16) translocation that has been mapped to the MAF oncogene at 16q23 near FRA16D (Krummel et al., 2000), and deletions within FRA16D have been found in cell lines derived from adenocarcinomas of the colon, breast, lung, stomach and ovary (O’Keefe and Richards, 2006). The two most highly expressed common fragile sites, FRA3B and FRA16D, are located within a tumor suppressor gene, FHIT for 3B and WWOX/FOR for 16D, and putative tumor suppressor genes are also found in several other fragile site regions (Huebner and Croce, 2001). Thus the deletion or rearrangement of tumor suppressor genes associated with common fragile sites may play a role in cancer development. This idea is supported by the result that ectopic WWOX expression inhibits tumor growth of breast cancer cells in a mouse model and inhibits anchorage-independent growth of breast cancer cell lines (Bednarek et al., 2001).
Thirteen common fragile sites have been cloned and characterized (Schwartz et al., 2006). In contrast to rare fragile sites, which are caused by expansion of a minisatellite or trinucleotide repeat, common fragile sites extend over hundreds of kilobases and do not appear to contain expanded repetitive elements. Using a computer program (FlexStab or TwistFlex) that predicts the flexibility of the DNA helix by using measurements of the twist angle between consecutive base pairs, the Kerem group showed that common fragile site regions contain clusters of flexibility peaks that are extremely AT rich (78% versus 61% for nonflexible regions) (Mishmar et al., 1998; Zlotorynski, 2003). The same computer analysis of FRA16D revealed a cluster of 6 flexibility peaks within the ~270 kb region defined as most fragile (Ried et al., 2000). Furthermore, the alignment of the flexibility peaks within FRA16D with two tumor cell lines revealed that several of the peaks were located near either a mapped deletion breakpoint or inside a deleted region (Ried et al., 2000). These results all suggest that the flexible regions are good candidates for determinants of fragility, but there has been no direct experimental evidence to support this model.
Common fragile sites have been shown to be late replicating regions (see Freudenreich, 2005 for review). For example, FRA3B replicates late in S phase and exposure to aphidicolin further delays replication (Le Beau et al., 1998), and FRA16D initiates late in S phase coupled with slow replication progress to G2 phase (Palakodeti et al., 2004). The expanded CGG repeats found at late replicating rare fragile site FRAXA have been shown to form hairpin and quadruplex structures that can stall a replication fork in bacteria or yeast cells (Mirkin, 2006). Thus secondary structures that stall or slow progression of the replication fork could cause the expression of fragile sites. The finding that cells deficient for ATR, which responds to blocked replication forks, exhibit increased expression of FRA3B, FRA16D and FRA7H further supports this model (Casper et al., 2002). In S. cerevisiae, cells mutant for the ATR homolog Mec1 exhibit increased chromosomal fragmentation at regions identified as replication slow zones (Cha and Kleckner, 2002), and deficiency or mutation of checkpoint proteins Mec1, Rad9, Mrc1 (human Claspin) or Rad53 (human Chk2) increases fragility of an expanded CAG/CTG repeat tract that acts as a fragile site in yeast (Lahiri et al., 2004; Freudenreich and Lahiri, 2004).
Despite evidence that supports the hypothesis that human common fragile site breakage is caused by either slow replication fork progression or replication fork barriers, it has never been directly proven. In addition, it is not known which sequences with the large fragile regions (100’s of kilobases) are responsible, and whether they inhibit replication by forming a DNA structure or by other means such as protein binding or collision with transcription. We have developed an assay to study human common fragile site FRA16D in yeast and used it to test whether regions of high flexibility increase chromosome breakage in a quantitative manner. We report that a short ~500 bp AT rich region that spans the peak of highest flexibility and contains a polymorphic perfect AT repeat increases chromosome fragility, whereas three other tested sequences within FRA16D do not. This is the first molecular determination of a sequence element within a common fragile site that increases chromosome breakage. In addition, we go on to show by two dimensional (2-D) gel analysis of replication intermediates that this sequence stalls a replication fork in a manner dependent on the length of the perfect AT repeat. The AT length that results in fork stalling correlates with predicted ability to form a cruciform structure. Our results support previous suggestions that replication fork stalling at a secondary structure can lead to the chromosome breakage observed at human common fragile sites.
RESULTS
A Genetic Assay to Study Fragility of FRA16D in Yeast
We sought to develop a genetic assay utilizing yeast that would allow a quantitative measure of the amount of breakage of common fragile site sequences or sub-regions. To this end, we modified an assay that our laboratory has previously used to analyze breakage of an expanded CAG/CTG repeat sequence (Callahan et al., 2003; Liu et al., 2004; Lahiri et al., 2004). This assay utilizes a yeast artificial chromosome (YAC) containing selectable markers at each end and a telomeric seed sequence (C4A4) to rescue broken YACs (Figure 1A). YACs were built that contained either very large (megabase) regions of human sequence that included the entire FRA16D sequence (Figure 1B) or small subregions of FRA16D (Figure 2A, 2B). If breaks occur in the FRA16D region, some portion of breakage intermediates will be degraded by exonucleases to expose the C4A4 sequence which is recognized by telomerase, resulting in rescue of the broken YAC by de novo telomere addition (Figure 1A). In this process cells are converted from 5-floroorotic acid sensitive (FOAS) to FOA-resistant (FOAR) due to the loss of the URA3 gene, and thus can be positively selected for in a fluctuation analysis to arrive at a rate of FOAR that correlates with a rate of breakage. In this paper we will use the term “fragile” to indicate a DNA sequence prone to breakage as assessed by cytogenetics, this genetic assay, or direct detection of chromosome breaks by gel electrophoresis.
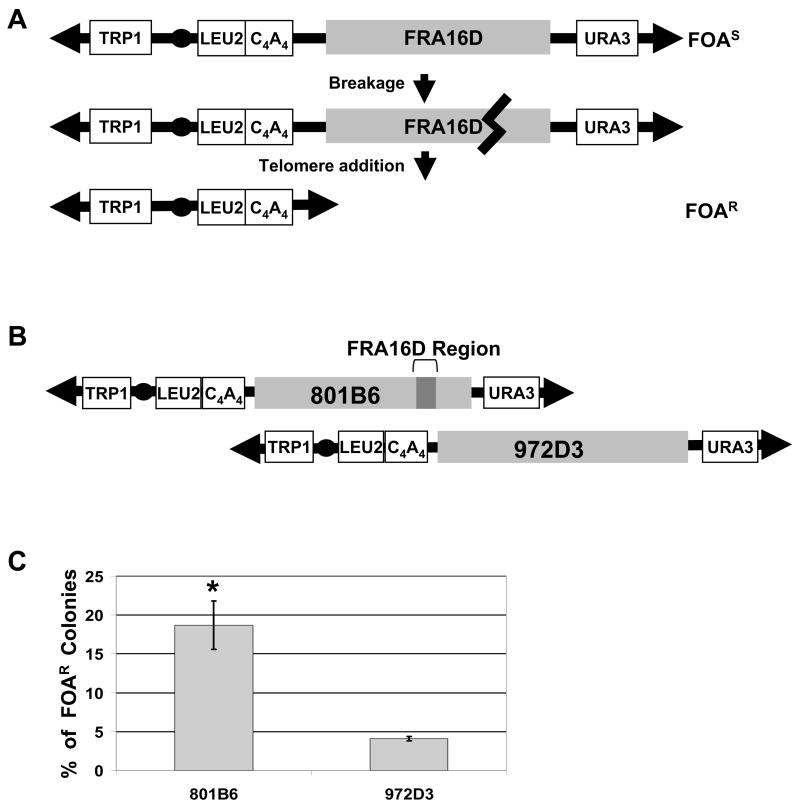
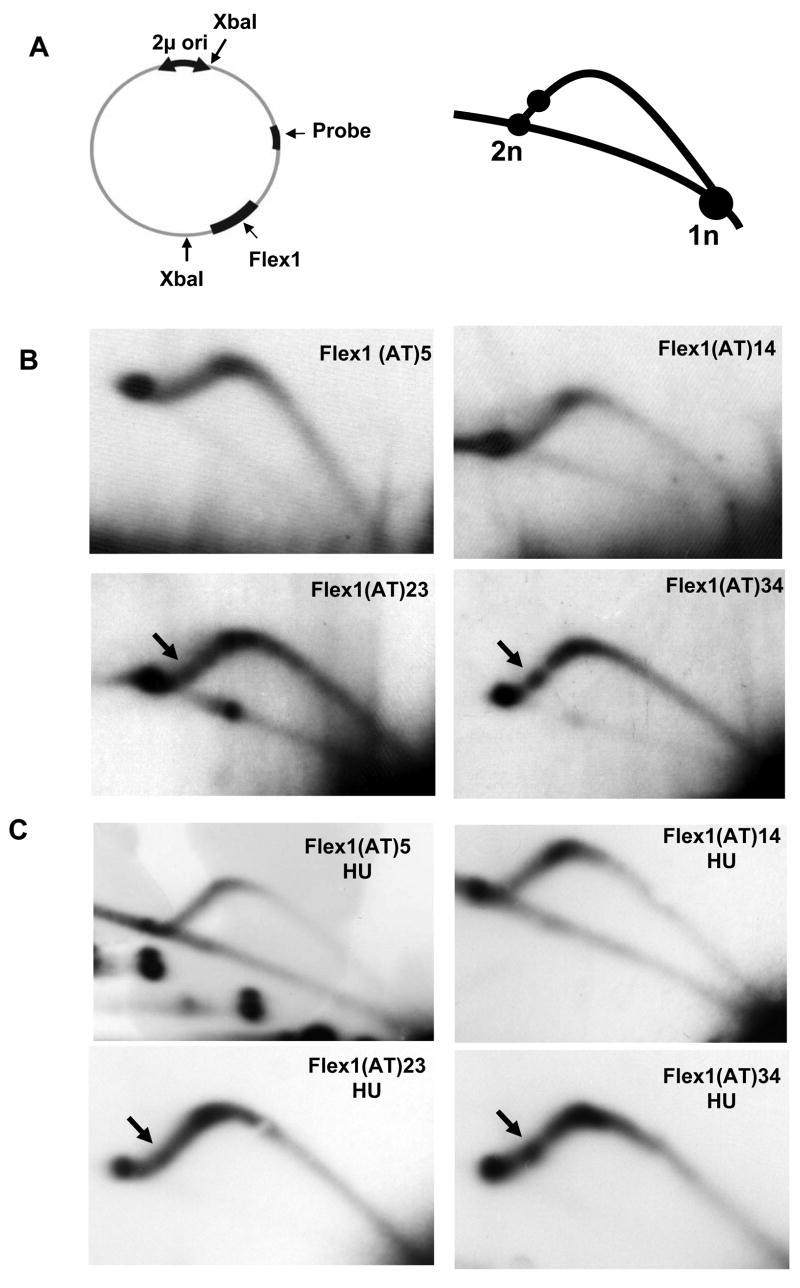
Figure 1. Assays to quantify FRA16D fragility and map sites of breakage.
(A) The YAC breakage assay. Cells containing a YAC, which contains either the entire FRA16D region or subregions and the URA3 gene, are FOAS. When breakage occurs inside the FRA16D region (heavy black line), the broken YAC can be rescued by the addition of a new telomere to the 108 bp C4A4 telomere seed sequence. The resulting cells will be Leu+, Trp+ and FOAR. (B) YACs from the CEPH YAC library were modified by adding a telomere seed sequence and a LEU2 marker. Human sequences are represented by grey boxes (not to scale). The dark grey box represents the 270 kb FRA16D region defined as most fragile by (Reid et al., 2000). YAC 801B6 and YAC 972D3 are aligned according to their coordinates in the human genome. (C) The breakage assay was performed for YAC 801B6 and YAC 972D3. Bars represent the average of 3 experiments, SEM is shown. Statistical significance was determined using a pooled variant t-test, * P < 0.05. (D) Breakage intermediates initiate within FRA16D. Cells of the rad50Δ strain background containing the 801B6 YAC from the CEPH YAC library were arrested in G1, released into S phase, and samples collected every 30 min for 3 hours. Chromosomes were prepared and separated using PFGE, blotted, and hybridized to either a TRP1 or a URA3 probe to the left or right arm of the YACs, respectively. The endogenous chromosome IV (Containing TRP1) and V (containing URA3) and full length YAC 801B6 are indicated by long arrows. Initial breakage intermediates are indicated by asterisks (** or *), degradation products by black arrowheads, putative recombination products by grey arrowheads. (E) Mapping of the breakage intermediates, symbols as in (D). The diagram of YAC 801B6 is approximately to scale.
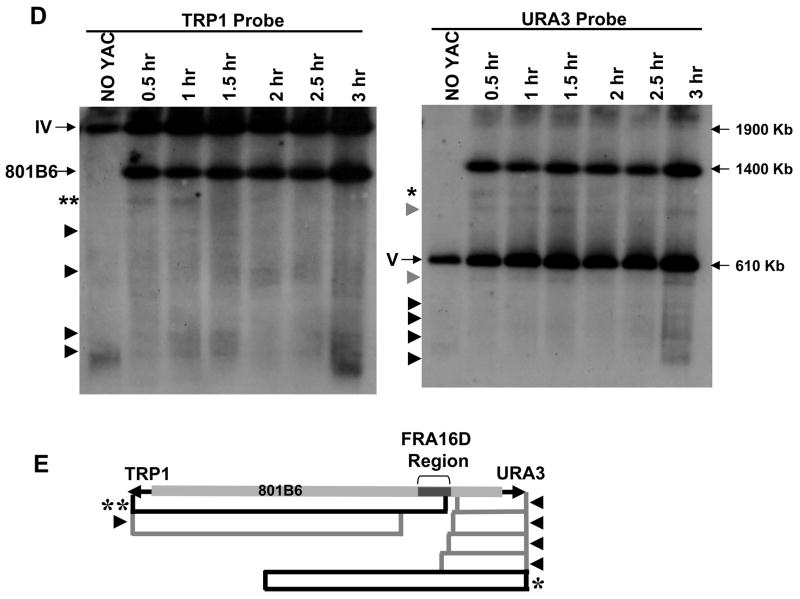
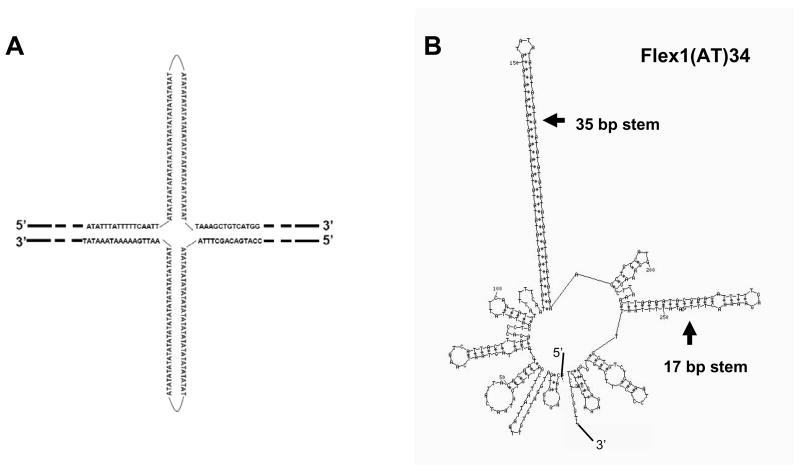
Figure 2. The Flex1 sequence increases chromosome breakage.
(A) Structure of the YACs containing the subregions of FRA16D diagrammed in (B) (denoted as “Flex”). Two types of YACs were created, either without (top) or with (bottom) the HIS5 gene. The black bars flanking the Flex region represent homologous sequences. For the four Flex1 sequences diagrammed, the dark grey bars represent the perfect AT repeat region. Flex1(AT)5, (AT)14 and (AT)23 are exactly the same except for the AT repeat number; Flex1(AT)34 contains 101 bp of extra distal sequence, and lacks 400 bp of the proximal sequences compared to the other Flex1 sequences. (B) The FlexStab score of the FRA16D region. Basepairs are on the X axis. Grey arrows represent the flexibility peaks used for this study. Below it, the exons of the WWOX/FOR gene are represented by grey boxes. Regions chosen for analysis in the present study are represented by black boxes: Flex4 is 1349 bp, Flex5-p is 2054 bp, Flex1 is 314–552 bp. The control sequence is 1320 bp or 400 bp. (C) The flexibility score of the right arm of the top YAC shown in (A). (D) Results of the breakage assay in the rad52Δ background using the top (URA3) YACs diagrammed in (A). The control is 1320 bp. The experiment was repeated at least 3 times for each strain, average with SEM is shown. * P < 0.05 compared to the control by a pooled variant t-test (E) Breakage assay using the Flex1-HIS5 YAC in the wild-type background in the absence (dark grey) or presence (light grey) of hydroxyurea. FOAR colonies were replica plated to YC-His plates, and only the His− colonies, indicating FOAR due to YAC breakage, were used to calculate a rate of FOAR His−. A 400 bp control was used (similar in size to the cloned Flex1 regions). The experiment was repeated at least 3 times for each strain, average with SEM is shown. * P < 0.05 compared to the respective control (either no drug control or 0.15 M hydroxyurea control) by a pooled variant t-test.
FRA16D Sequences Break in vivo Even in the Absence of a Replication Inhibitor
To quantify the FRA16D breakage rate, we modified two YACs from the CEPH YAC library for use in the genetic assay. The 801B6 YAC contains 1.4 Mb of human sequence including the 270 kb FRA16D region that contains six flexibility peaks (see Fig. 2B), and the 972D3 YAC contains 1.6 Mb of human sequence distal to FRA16D (Figure 1B). Results show that cells with the FRA16D-containing YAC (801B6) exhibited a significant 4.6-fold increase of FOAR compared to cells with the control 972D3 YAC (18.7% versus 4.1%) even in the absence of any replication inhibitor (Figure 1C). An estimation of the rate of breakage is 4.4 × 10−6 per kb for the control 972D3 YAC, and 20 × 10−6 per kb for YAC 801B6. Physical analysis of YACs from some of the FOAR colonies showed that they were shorter than the original YAC, consistent with a breakage event (Supplemental data). FlexStab analysis of both YACs did not show a significant difference in the number of peaks of score 13.7 or above (as defined by Zlotorynski, 2003), as 801B6 had an average of 1 peak per 38 kb and 972D3 showed ~1 peak per 41 kb. One difference is that YAC 801B6 had more peaks of score ≥ 16 than 972D3 (5 vs. 3) and contained the three highest peaks (highest =18.2 at Flex1 (Fig. 2B); 16.9, 16.6).
To determine the location of the breaks, we used a physical assay (PFGE) to directly detect breakage intermediates. One or more preferred fragile regions were observed in the 801B6 YAC compared to the 972D6 YAC (Figure S1). To more clearly define where breaks initiate within the 801B6 YAC, we synchronized cells in G1, released into S phase, and performed a time course experiment (Figure 1D). To facilitate detection of broken intermediates, the experiment was done in a strain lacking the Rad50 protein (rad50Δ) which has been shown to exhibit very slow degradation of broken ends. The largest initial breakage intermediate visualized by the TRP1 probe (**) maps to the FRA16D region containing the six flexible peaks, providing direct evidence that this is a preferred site of chromosome breakage in vivo, even in the absence of aphidicolin (Figure 1 D,E). At later time points, this band disappeared and smaller bands appeared that we interpret as degradation products (the smallest bands are likely caused by mechanical force during sample preparation, since they could be observed in control cells without the YAC). We were not able to observe an early breakage product that mapped to FRA16D with the URA3 probe, but the smaller bands appearing at later time points were consistent with FRA16D being the most preferred site of breakage (Figure 1E). The URA3 probe did, however, reveal an early breakage product that maps ~580 kb proximal to FRA16D, which might represent another hot spot for breakage. The bands indicated by grey arrows are likely stable recombination products generated during growth before cells were synchronized, since they existed at the beginning and persisted throughout the experiment. In contrast, the same time course using cells containing the control 972D3 YAC showed no specific breakage intermediates (Figure S1). In summary, these results show direct molecular evidence that there are intrinsic elements both within the FRA16D region and perhaps also ~580 kb proximal to this region that cause chromosomes to break, and that these elements are fragile in the context of a yeast cell and in the absence of a replication inhibitor.
The Flex1 High Flexibility Sequence Causes Chromosome Fragility
In order to directly test whether the sequences at or near the high flexibility peaks can cause increased chromosome breakage, we inserted several short regions of FRA16D into a YAC that contains all the elements necessary for the genetic breakage assay (Figure 2A). We chose three AT-rich flexibility peaks and/or surrounding sequences and a control sequence also located inside FRA16D but not spanning any of the flexibility peaks (Figure 2B). Flex4 is 1349 bp and includes the Flex4 peak sequence, an (AT)8 perfect repeat, and sequences that map to an AGS tumor cell line breakpoint (Figure 2B; Ried et al., 2000). The 2054 bp Flex5-proximal (Flex5-p) sequence is very close to Flex5 but does not include the actual Flex5 peak sequence, and maps to a breakpoint in the AGS cell line. Flex1 is ~500 bp and contains the Flex1 peak sequences and sequences flanking a breakpoint in the HCT116 tumor cell line. All the sequences are AT rich: 55–58% for Flex4 and Flex5-p; 65–75% for Flex1. Flex1 also contains a perfect AT/TA di-nucleotide repeat within the AT-rich sequence that shows a high degree of polymorphism of repeat number in the population (Finnis et al., 2005), therefore we inserted 4 different Flex1 sequences with different numbers of AT repeats (AT-5, -14, -23 or -34; Figure 2A).
To verify that the human flexibility peaks manifest as peaks in the context of the YAC and in a yeast background, we performed a flexibility analysis on the right arm of the YAC containing Flex1(AT)34 and on a region of yeast chromosome I (Figure 2C, Figure S2). Flex1(AT)34 gave a peak of ~17, similar to the score of 18 shown by Reid et al. using an (AT)55 allele, and the background flexibility score of the YAC and the yeast chromosome (score of 11) is similar to the FRA16D region. Thus the Flex YACs mimic the circumstances of the endogenous FRA16D region in terms of predicted sequence flexibility.
We used the genetic assay to test whether high flexibility peaks increase chromosome fragility. Cells that contained Flex1(AT)5, Flex1(AT)14 and Flex1(AT)23 all showed a significantly increased rate of FOAR compared to the control sequence in a recombination-deficient rad52Δ background, whereas cells containing Flex4 and Flex5-p did not (Figure 2D; note that some homology flanking the Flex regions precluded accurate quantification in WT strains for these YACs, see Supplemental Data). Although not significant (p=0.05–0.18), the rate of FOAR of Flex1-(AT)23 (5.8-fold over control) was consistently greater than Flex1-(AT)5 and Flex1-(AT)14, suggesting that AT repeat length may be a factor in the propensity to break. Interestingly, the negative result for the Flex5-p sequence which includes the region mapped as a breakpoint in the AGS tumor cell line suggests that the breakpoint reflects how a double strand break was healed, rather than indicating a hot spot for breakage. YACs from a subset of FOAR colonies were characterized molecularly by Southern blot analysis to detect the site of healing. The majority had healed at the C4A4 site as predicted by a breakage event in the Flex1 sequence (Supplemental Data).
In order to test the fragility of Flex1 in a wild-type background, we replaced one of the homologous sequences with a HIS5 marker (Figure 2A). The addition of the HIS5 marker also allowed us to subtract out events that caused FOAR for reasons other than YAC breakage (Figure 2E). In yeast, hydroxyurea can be used to efficiently slow or stop replication, analogous to the conditions used to induce expression of common fragile sites in humans (although aphidicolin is a more effective and specific inducer of common fragile sites than hydroxyurea in human cells, yeast replicative polymerases are 3000–4000x more resistant to aphidicolin (Cooley and Mishra, 2000) and we were not able to achieve a condition that effectively inhibited replication). Wild-type cells that contained Flex1(AT)14 and Flex1(AT)23 showed a significant increase in the rate of YAC breakage compared to the control sequence in both the presence or absence of hydroxyurea (Figure 2E; control rate = 3.1 × 10−6 per kb). Cells that contained Flex1(AT)5 showed a significant increase of FOAR only in the presence of hydroxyurea. Altogether, these results show that the Flex1 region is inherently fragile, and deficiency in homologous recombination or replication stress can increase the level of fragility.
Unlike the other Flex1 sequences, Flex1(AT)34 showed a rate of FOAR similar to or significantly lower than the control in both the rad52Δ and wild-type backgrounds (Figure 2D, 2E). One explanation for this result is that the (AT)34 repeat somehow interfered with recovery of broken YACs, for example by interfering with telomere addition. In addition to YAC breakage, FOAR can arise by URA3 point mutation (rate ~10−8), or by mutations that impair FOA import or metabolism. Poor recovery of broken YACs by telomere addition will lead to a greater proportion of FOAR colonies that are due to one of these alternative events and contain a full-length YAC. To test the proportion of intact vs. broken YACs, FOAR colonies were replica plated onto YC-His plates. FOAR cells containing the control region showed a His− frequency of 76% (3287/4312), whereas cells containing Flex1(AT)23 showed 86% (5644/6562) His− colonies, consistent with the increased rate of breakage observed. Strikingly, Flex1(AT)34 -containing cells exhibited only 47% (493/1045) His− colonies, which fits the prediction of poor recovery of broken YACs. When DNA from some of the FOAR colonies was characterized molecularly, all FOAR His+ colonies analyzed contained a full-length unbroken YAC (10 of 10), whereas all FOAR His− colonies contained a YAC that had healed at the C4A4 telomere seed sequence.
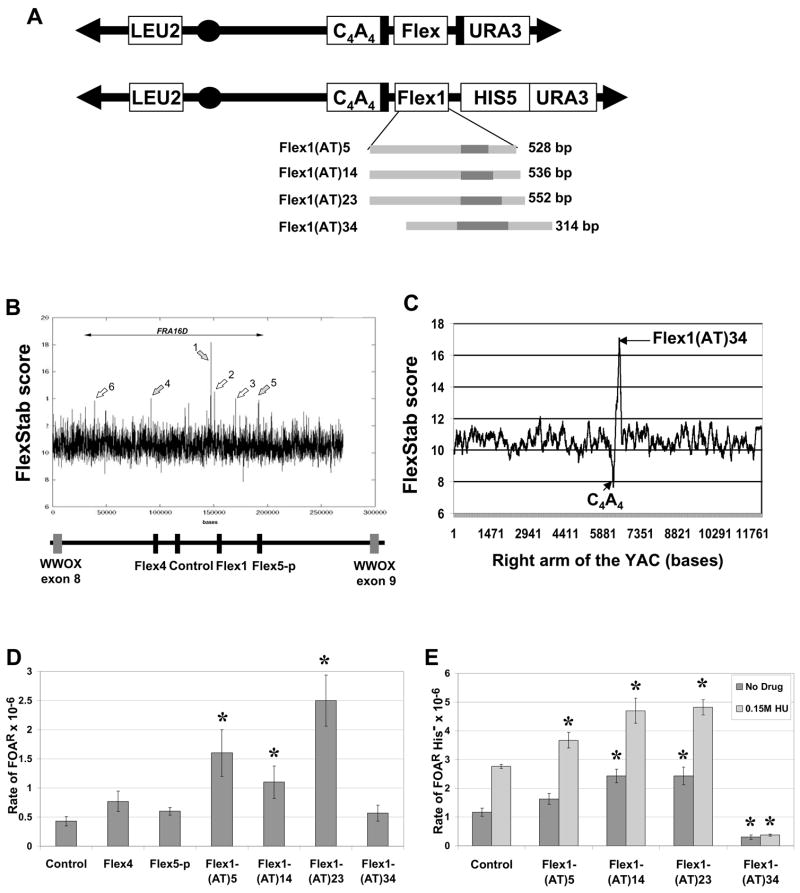
Interestingly, repeats of (AT)21–34 in the context of an AT-rich sequence have been shown to form a cruciform structure in vivo in E. coli, whereas lower numbers of AT repeats do so less efficiently (McClellan et al., 1990; Dayn et al., 1991). Therefore, the (AT)23 and (AT)34 alleles of the Flex1 sequence could potentially also form a cruciform in vivo under the right conditions (Figure 3A). The Flex1 region is also predicted to form strong secondary structures by the Mfold program (Zuker, 2003) (Figure 3B). The (AT)34 perfect repeat forms a predicted hairpin with a melting temperature (Tm) of 67°C, as well as a predicted 17 bp hairpin (Tm 52.7°C) 23 bp away that is not present in the other three Flex1 sequences tested (in these the cloned region ends 24 bp from the variable length AT hairpin). Agarose and polyacrylamide gel analysis of PCR products of each Flex1 allele strongly suggested that the Flex1 (AT)23 and (AT)34 alleles formed secondary structures, with the Flex1(AT)34 DNA forming a structure a higher percentage of the time (data not shown), and our 2D-gel data (see below) is also consistent with this idea. Thus, we hypothesize that the Flex1(AT)34 sequence may actually break at a high rate, but forms a DNA structure that inhibits de novo telomere addition and recovery in our assay, for example by inhibiting exonuclease degradation of the end of the break needed to expose the C4A4 telomere seed. If this hypothesis is correct, the Flex1(AT)23 breakage rate may also be an underestimate and the length of the AT repeat and its propensity for secondary structure formation may be a contributing factor in determining the level of fragility.
Figure 3. Flex1(AT)34 is predicted to form a strong secondary structure.
(A) Putative cruciform structure of Flex1(AT)34. (B) The secondary structure of the 314 bp fragment containing Flex1(AT)34 predicted by the Mfold Program (highest stability structure, Tm 60 °C). http://www.bioinfo.rpi.edu/applications/mfold/dna/ (Zuker, 2003).
The Flex1 Region Causes Site-Specific Replication Fork Stalling
The results that the Flex1 sequence increased chromosomal breakage and that breakage was further exacerbated by hydroxyurea suggested that it could be a replication fork barrier. To determine directly whether the Flex1 region can stall or slow replication, we analyzed replication intermediates by two-dimensional gel electrophoresis (2D-gel). Flex1 regions with the AT repeat numbers used in the fragility assay were cloned into a plasmid containing a 2 micron (2μ)replication origin (Figure 4A). Yeast cells transformed with these plasmids were grown to early log phase and DNA prepared for 2D-gel analysis. If Flex1 sequences cause replication fork stalling, we would expect to observe an accumulation of replication intermediates which will be visible as a bulge on the short arm of the arc (Figure 4A). 2D-gel analysis revealed a strong bulge that maps to the Flex1 sequence containing (AT)34 (Figure 4B). For Flex1(AT)23, a fainter bulge was observed that mapped to the same location (Figure 4B). Smooth replication arcs were observed for fragments containing Flex1(AT)5 and Flex1(AT)14 and the plasmid control (Figure 4B and data not shown). The same pattern was seen for cells grown in hydroxyurea, and the drug did not visibly affect the strength or appearance of the stalls (Figure 4C). It is possible that any additional amount of stalling in hydroxyurea treated cells is offset by the increased fragility of the sequence and resulting failure to recover intact replication intermediates. Altogether, our data demonstrate that the Flex1 sequence can efficiently stall a replication fork in a manner dependent on the length of the AT repeat. Correlation with predictions of secondary structure suggests that a structure formed by the AT repeat, potentially in combination with surrounding structures, is responsible for the fork stalling.
Figure 4.
Flex1(AT)34 can stall replication fork progression. (A) Diagram of the plasmid containing the Flex1 regions used for 2D-gel analysis and predicted position on the Y arc of replication intermediates of a stall within Flex1 (circle). The 2μ origin replicates bi-directionally. (B) 2D-gel analysis of the replication intermediates traversing the 2914 bp XbaI fragment from plasmids replicated in yeast and containing the indicated Flex1 region. The AT perfect repeat is 191 bp from the second XbaI site for (AT)34 and 97 bp for (AT) 23, (AT)14 and (AT)5. A second experiment yielded the same results (not shown). (C) 2D gel analysis of replication intermediates isolated from cells grown in 0.1 M hydroxyurea (HU).
The FRA16D Region Likely Contains One or More Replication Fork Barriers in a Chromosomal Context
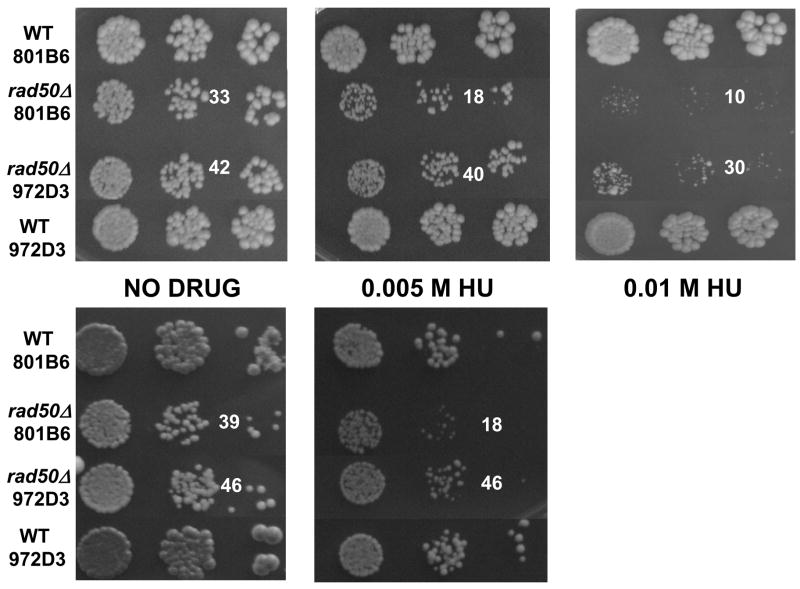
RAD50 is a member of the MRX (MRN in mammals) complex that senses and binds to DSBs to facilitate repair, including DSBs that arise during replication (D’Amours and Jackson, 2002). It has also been shown that the MRN complex localizes to restarting forks and that RAD50 is required for cell viability in response to replication fork stalling (Trenz et al., 2006; Lambert et al., 2005). To assess whether Flex1 or other sequences within FRA16D might stall forks within a chromosomal context with a great enough frequency to cause a phenotypic effect, we performed a viability assay by spotting a serial dilution of cells containing the FRA16D 801B6 YAC or the control 972D3 YAC. If replication barriers exist within the FRA16D region, we should observe poor growth of rad50Δ cells containing that sequence. An equal number of cells of each strain were plated in the presence and absence of hydroxyurea. On plates without hydroxyurea, fewer cells containing the 801B6 YAC produced a colony, indicating a slight growth disadvantage compared to cells containing the 972D3 YAC in a rad50Δ background (Figure 5, 2 experiments shown). When exposed to hydroxyurea, cells containing the 801B6 YAC exhibited a marked decrease in both size and number of colonies compared to cells containing the 972D3 YAC. The growth difference was exacerbated with increasing amounts of hydroxyurea (Figure 5). No growth difference was detected in wild-type strains. Therefore the combination of hydroxyurea and a rad50Δ resulted in a significant growth defect in cells containing the FRA16D sequence compared to the control sequence. These data suggest that there are replication fork barriers in the FRA16D region in vivo and that Rad50 is needed to restart these stalled or collapsed replication forks and/or repair resulting breaks.
Figure 5.
FRA16D-dependent growth defect in the rad50Δ background. An equal amount cells containing either YAC 801B6 or YAC 972D3 were spotted on YC-Trp-Ura plates containing either no drug or 0.005 M or 0.01 M hydroxyurea. Three dilutions of 10−1, 10−2 and 10−3 are shown; the number of colonies in the second dilution is indicated. The top three panels represent one experiment; the bottom two panels represent a separate, independent experiment. Each panel shows colonies from one plate, all plates were incubated 3 days at 30 °C.
DISCUSSION
It has long been proposed that the molecular basis for fragility at both common and rare fragile sites is slowed replication (Laird, 1987; Le Beau et al., 1998; Zlotorynski et al., 2003; Glover et al., 2005; Haber and Debatisse, 2006). However until now, this hypothesis has not been directly tested in vivo. In this study we show that a ~500 bp region encompassing Flex1, an AT-rich region predicted to have high flexibility within common fragile site FRA16D, is a hot spot for breakage in yeast. Furthermore, we have demonstrated that the Flex1 region stalls replication fork progression in a manner dependent on the length of a variable perfect AT repeat predicted to form a cruciform structure. Therefore, replication fork stalling at a secondary structure within the Flex1 sequence could account, as least in part, for the increased breakage observed in the FRA16D region. By extension, other human common fragile sites which contain similar structure-forming regions may break via the same type of mechanism.
Fork Stalling and Fragility of Flex1 may be due to Structure Formation by the Perfect AT Repeat Region in the Context of the Flex1 AT-Rich Sequence
The Flex1 sequence is highly AT rich (65–75%) and Flex1 single strands are predicted to form multiple hairpin structures (Fig 3A). In addition, studies in vitro and in vivo have shown that AT-rich inverted repeats easily form a cruciform structure involving both strands (Fig 3B) because AT repeats have fast relaxation kinetics which allows melting of the double helix (Bowater et al., 1991; Dayn et al., 1991). Helix melting and cruciform formation can be achieved in vivo by any process that increases DNA supercoiling such as active transcription, replication, or blockage of protein synthesis and it depends on the length of the inverted repeat and also on the AT content of the flanking sequences (McClellan et al., 1990; Dayn et al., 1991). Because the replication fork stall was so strong for the (AT)34 tract in the context of Flex1, we favor the idea that a cruciform involving both strands is forming, rather than a hairpin on just one strand (Figure 3B). Supporting this idea, much longer CTG or CAG repeat sequences, such as (CTG)80, which thermodynamically should form a more stable hairpin, do not detectibly stall a replication fork in yeast cells (Pelletier et al., 2003). In addition, long CTG/CAG sequences do not inhibit telomere formation in our YAC breakage assay whereas the Flex1(AT)34 sequence likely did, suggesting that the structure formed on the chromosome in vivo may be fundamentally different from a hairpin.
The AT-rich context of Flex1 likely plays an important role in facilitating structure formation. For example, structure formation at the perfect AT repeat might nucleate unwinding of the surrounding AT-rich sequence to allow the formation of multiple hairpins. One of these may be the shorter predicted 17 bp hairpin present in the Flex1(AT)34 clone, which could form a second structure that strengthens the fork stalling we observed. Another reason to emphasize the context of the AT repeat is that Flex1(AT)5 and (AT)14, which did not show detectable fork stalling by 2D gel analysis (although low levels would not be detected by this technique) did exhibit significant increases in fragility, whereas the Flex4 (AT)8 sequence did not. In summary, we favor the idea that the fork stalling we observed is due to cruciform formation by the perfect AT repeat region in the context of the AT-rich Flex1 sequence, and that inhibition of replication contributes significantly to the fragility of the Flex1 sequence.
Contribution of Flex1 to the Fragility of the FRA16D Region
In the time course experiment (Figure 1D), we observed that double-strand breaks initiate in the FRA16D region, demonstrating that this region does accumulate double strand breaks at the molecular level, consistent with cytogenetic and in vivo data in humans. Although not directly comparable because the efficiency of recovery of broken YACs may differ, the Flex1(AT)23 sequence broke 2.1-fold over the control (no drug, WT background), whereas the YAC containing the entire FRA16D region had a frequency of FOAR 4.6-fold over the control under the same conditions, suggesting that Flex1 is a major, but perhaps not the only, determinant of fragility. Supporting an important role for Flex1, all the tumor cell lines with deletions in FRA16D that have been studied in detail have the Flex1 sequences either deleted or at the boundary of the deletion, and in fact one breakpoint mapped within the Flex1 AT repeat (Finnis et al., 2005). Flex1 also has a much higher flexibility score (18.2) compared to the other peaks of flexibility such as Flex4 (score 14), and in fact is the highest peak in the entire 3 Mb of the 801B6 and 972D3 YACs (the next highest peak score is 16.9). Nonetheless, the high frequency of FRA16D expression could indicate that other sequences that inhibit replication fork progression may also exist, which would act synergistically to increase the probability of replication fork stalling over a large region. For example other peaks identified by the FlexStab program, small inverted repeats mapped by Ried et. al. (2000), or the breakage site we identified ~580 kb proximal to FRA16D could contribute to the overall fragility of the region under conditions of replication stress. Based on the results with the different Flex1 sequences and the negative result with Flex4, we hypothesize that the most important determinant of fragility is the ability to stall replication, and that sequences with this capacity will often, but not always, overlap with flexibility peaks because the ability to easily unwind can allow a DNA structure to form.
A Model for the Mechanism behind the Chromosome Breaks and Rearrangements Found at Common Fragile Sites
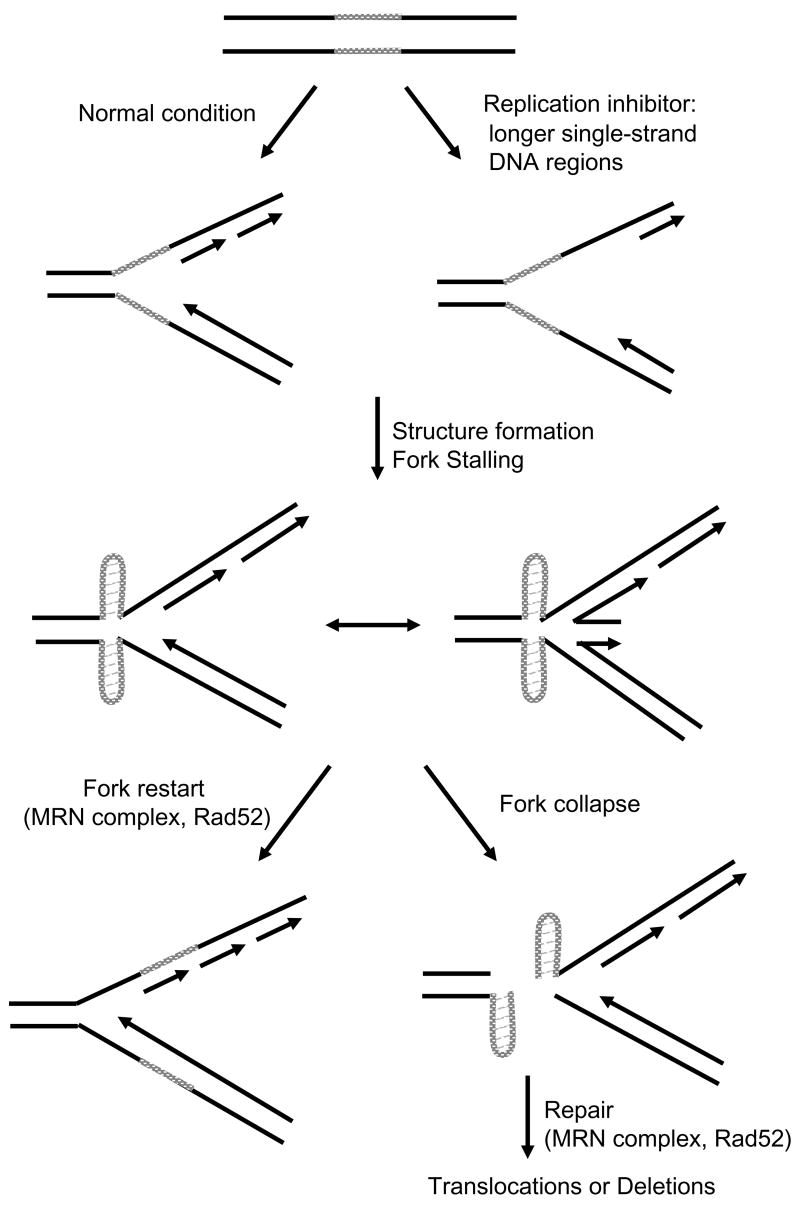
Our data are consistent with a model in which the Flex1 region causes chromosome breakage by forming one or more secondary structures, such as a cruciform or multiple hairpins that can stall a replication fork (Figure 6). A stalled fork could collapse to cause chromosome breakage or initiate recombination during a fork restart attempt. In human cells, common fragile sites are not expressed at a detectable level unless induced by aphidicolin, an inhibitor of polymerase α, δ and ε, at a concentration that slows but does not stop replication (Glover et al., 2005). This condition leads to more single-stranded DNA at the replication fork which could facilitate the formation of a cruciform or hairpin structure by the AT perfect repeats (Figure 6). Once the structure forms, it will stall the replication fork, requiring checkpoint proteins (such as ATR) and repair pathways (such as Rad52-dependent strand invasion) to facilitate fork restart. If fork restart fails, replication fork collapse will result in a DNA break (Figure 6). Mutations that increase expression of fragile sites, such as the ATR pathway, may do so not only by regulating fork progression and/or restart, but also by increasing the single-stranded DNA region at the fork to facilitate structure formation. It is interesting that cancer cells with deletions at fragile sites often originate from tissues such as the lung and GI tract that are either highly proliferative or are more frequently exposed to chemicals that could cause replication fork stress, such as caffeine, ethanol, and nicotine (Richards, 2001).
Figure 6.
A Model for the Mechanism behind the Chromosome Breaks and Rearrangements Found at Common Fragile Sites. The type of secondary structure and exact location with respect to the stalled fork and broken chromosome are unknown, but are illustrated for diagrammatic purposes.
Since the efficiency of fork stalling was dependent on the length of the Flex1 perfect AT repeat, human alleles with longer AT repeats could be more fragile. Interestingly, Finnis et al (2005) found that the Flex1 AT repeat is highly polymorphic in the population (97% heterozygosity) with alleles varying between 11–88 AT repeats, and alleles of 34 repeats or greater are quite common. Although a relationship between AT length and fragility could not be documented in the Finnis et al. study, a comparison of fragility rates of the different alleles in our assay suggests that the effect may be subtle. Since the longer (AT)34 allele also interfered with our assay to prevent quantification, it will be important to develop alternative methods to rigorously test this hypothesis.
The Consequences of Fork Stalling in the FRA16D Flex1 Region
We observed increased Flex1 breakage in a rad52Δ background, and increased cell lethality in rad50Δ cells containing the FRA16D region, indicating that both the Rad52 and Rad50 proteins are important for preventing Flex1/FRA16D breakage. This result is consistent with data showing that Rad51 downregulation exacerbates FRA16D and FRA3B fragility and that a Rad50 complex member, DNA-PKcs, forms foci at FRA3B in human cells treated with aphidicolin (Schwartz et al., 2005). The MRN complex containing both Mre11 and Rad50 is recruited to collapsed forks by ATR/ATM and plays a role in fork restart (Trenz et al., 2006). Either Rad52-dependent strand invasion or MRN could facilitate fork restart or repair a break induced by a fork collapse, which could explain why these proteins are important in preventing Flex1 breakage and promoting survival of cells containing a fragile site (Figure 6).
Chromosomal instability, such as deletions and translocations, can be an initial step for cancer development. Common fragile site regions undergo frequent chromosomal rearrangements that may play a role in tumorigenesis. In particular, the loss of tumor suppressor genes FHIT and WWOX, that span FRA3B and FRA16D respectively, has been observed in a large number of cancer cell lines (Huebner and Croce, 2001; Mangelsdorf et al., 2000). The instability of FRA16D occurs at the same location in various cancer cell lines, indicating that the instability of FRA16D may take place at an early stage and play a pivotal role in cancer development (Finnis et al., 2005). Our data suggest that the FRA16D Flex1 sequence, by virtue of stalling replication and causing chromosome breakage, may play a key role in the generation of the deletions and translocations found at FRA16D in cancer cells. The fact that longer Flex1 AT alleles caused a significantly stronger replication barrier predicts that human alleles with longer AT length at the Flex1 locus could predispose to cancer-causing rearrangements in the FRA16D region.
EXPERIMENTAL PROCEDURES
Yeast Strains and YACs
The 801B6 and 972D3 YACs (long YACs) are in the AB103 strain (wild-type or rad50Δ). YACs with various Flex sequences (short YACs) were introduced into a wild-type BY4705 strain or a rad52Δ background of the VPS105 strain by a kar cross (Callahan et al., 2003).
YAC Breakage Assay
All strains in all steps of all experiments were maintained on media that selected for presence of the YAC: YC-Trp-Leu for the long YACs, YC-Leu for the short YACs. Cells containing a YAC were grown to single colonies on –Ura selective plates. A fluctuation analysis was performed by separately growing ten colonies from each strain in 1 ml of liquid media (+Ura) for 6–7 doublings to allow breakage to occur, and plating a portion of each culture on media either containing or lacking FOA. For hydroxyurea experiments, drug was added to cells (OD 0.1–0.2) to a final concentration of 0.15 M. Cells were allowed to grow until OD 1–2, washed 3x in dH2O, resuspended in fresh YC-Leu media, and grow until OD 10. A rate of FOAR was calculated using the method of the median (Lea and Coulson, 1949). A subset of YACs from FOAR colonies were analyzed by Southern blot to map the size of the YAC and site of healing (see Supplemental data for details).
Analysis of Breakage by PFGE
Chromosome breakage intermediates were characterized using CHEF gels (BioRad) and Southern blot hybridization. To synchronize cells in G1, cells were grown to OD 0.4 in YEPD (pH 3.9), α-factor was added to a final concentration of 2.0 μg/ml, and cells were incubated 2 hours and monitored for arrest under the microscope. Cells were released to S phase by pelleting and washing twice in YEPD (pH 7.0). Aliquots were collected every 30 minutes. Chromosomes were prepared in plugs (Freudenreich et al., 1998) and separated in a 1% gel using a BioRad CHEF apparatus.
Analysis of Replication Intermediates
Flex1 sequences were cloned into both pRS426 and pRS406 plasmids at the EcoRI site and were transformed to the yeast BY4741 strain. XbaI fragments from pRS426 plasmids were used for 2D-gel analysis of replication stalling. For hydroxyurea experiments, a final concentration of 0.1 M was added to cultures at O.D.= 0.1–0.2. Cells were collected at ~O.D.= 1.0 and genomic DNA was purified, restriction enzyme digested, and separated on 2D-gels as in (Ivessa et al., 2002).
Supplementary Material
Acknowledgments
We especially thank Rob Richards and Merran Finnis for the cloned FRA16D sequences and for many helpful conversations related to this project. Thanks also to Sergei Mirkin for helpful suggestions and critical reading of the manuscript, to Alex Morgan, Arthur Brady and Lenore Cowen for help in running the program to predict DNA helix flexibility, Aviva Liebert and Phil Starks for use of their PAGE system, and Daniel Coleman for technical assistance. This work was supported by NIH grant GM63066 to C.H.F.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bednarek AK, Keck-Waggoner CL, Daniel RL, Laflin KJ, Bergsagel PL, Kiguchi K, Brenner AJ, Aldaz CM. WWOX, the FRA16D gene, behaves as a suppressor of tumor growth. Cancer Res. 2001;61:8068–8073. [PubMed] [Google Scholar]
- Bowater R, Aboul-ela F, Lilley DM. Large-scale stable opening of supercoiled DNA in response to temperature and supercoiling in (A + T)-rich regions that promote low-salt cruciform extrusion. Biochemistry. 1991;30:11495–11506. doi: 10.1021/bi00113a003. [DOI] [PubMed] [Google Scholar]
- Callahan JL, Andrews KJ, Zakian VA, Freudenreich CH. Mutations in yeast replication proteins that increase CAG/CTG expansions also increase repeat fragility. Mol Cell Biol. 2003;23:7849–7860. doi: 10.1128/MCB.23.21.7849-7860.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper AM, Nghiem P, Arlt MF, Glover TW. ATR Regulates Fragile Site Stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- Cha RS, Kleckner N. ATR Homolog Mec1 Promotes Fork Progression, Thus Averting Breaks in Replication Slow Zones. Science. 2002;297:602–606. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
- Cooley M, Mishra NC. Genetic analysis of the in vivo role of DNA polymerases in Saccharomyces cerevisiae. Curr Genet. 2000;38:256–263. doi: 10.1007/s002940000163. [DOI] [PubMed] [Google Scholar]
- Coquelle A, Toledo F, Stern S, Bieth A, Debatisse M. A new role for hypoxia in tumor progression: induction of fragile site triggering genomic rearrangements and formation of complex DMs and HSRs. Mol Cell. 1998;2:259–265. doi: 10.1016/s1097-2765(00)80137-9. [DOI] [PubMed] [Google Scholar]
- D’Amours D, Jackson SP. The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat Rev Mol Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- Dayn A, Malkhosyan S, Duzhy D, Lyamichev V, Panchenko Y, Mirkin S. Formation of (dA-dT)n cruciforms in Escherichia coli cells under different environmental conditions. J Bacteriol. 1991;173:2658–2664. doi: 10.1128/jb.173.8.2658-2664.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnis M, Dayan S, Hobson L, Chenevix-Trench G, Friend K, Ried K, Venter D, Woollatt E, Baker E, Richards RI. Common chromosomal fragile site FRA16D mutation in cancer cells. Hum Mol Genet. 2005;14:1341–1349. doi: 10.1093/hmg/ddi144. [DOI] [PubMed] [Google Scholar]
- Freudenreich CH. Molecular mechanisms of chromosome fragility. Chemtracts-biochemistry and molecular biology. 2005;18:141–152. [Google Scholar]
- Freudenreich CH, Kantrow SM, Zakian VA. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- Freudenreich CH, Lahiri M. Structure-Forming CAG/CTG Repeat Sequences are Sensitive to Breakage in the Absence of Mrc1 Checkpoint Function and S-phase Checkpoint Signaling: Implications for Trinucleotide Repeat Expansion Diseases. Cell Cycle. 2004;3 doi: 10.4161/cc.3.11.1246. [DOI] [PubMed] [Google Scholar]
- Glover TW, Arlt MF, Casper AM, Durkin SG. Mechanisms of common fragile site instability. Hum Mol Genet. 2005;14(Spec No 2):R197–205. doi: 10.1093/hmg/ddi265. [DOI] [PubMed] [Google Scholar]
- Glover TW, Stein CK. Induction of sister chromatid exchanges at common fragile sites. Am J Hum Genet. 1987;41:882–890. [PMC free article] [PubMed] [Google Scholar]
- Haber JE, Debatisse M. Gene Amplification: Yeast Takes a Turn. Cell. 2006;125:1237–1240. doi: 10.1016/j.cell.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Huebner K, Croce CM. FRA3B and other common fragile sites: the weakest links. Nat Rev Cancer. 2001;1:214–221. doi: 10.1038/35106058. [DOI] [PubMed] [Google Scholar]
- Ivessa AS, Zhou JQ, Schulz VP, Monson EK, Zakian VA. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummel KA, Roberts LR, Kawakami M, Glover TW, Smith DI. The characterization of the common fragile site FRA16D and its involvement in multiple myeloma translocations. Genomics. 2000;69:37–46. doi: 10.1006/geno.2000.6321. [DOI] [PubMed] [Google Scholar]
- Lahiri M, Gustafson TL, Majors ER, Freudenreich CH. Expanded CAG repeats activate the DNA damage checkpoint pathway. Mol Cell. 2004;15:287–293. doi: 10.1016/j.molcel.2004.06.034. [DOI] [PubMed] [Google Scholar]
- Laird CD. Proposed mechanism of inheritance and expression of the human fragile-X syndrome of mental retardation. Genetics. 1987;117:587–599. doi: 10.1093/genetics/117.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S, Watson A, Sheedy DM, Martin B, Carr AM. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005;121:689–702. doi: 10.1016/j.cell.2005.03.022. [DOI] [PubMed] [Google Scholar]
- Le Beau MM, Rassool FV, Neilly ME, Espinosa R, 3rd, Glover TW, Smith DI, McKeithan TW. Replication of a common fragile site, FRA3B, occurs late in S phase and is delayed further upon induction: implications for the mechanism of fragile site induction. Hum Mol Genet. 1998;7:755–761. doi: 10.1093/hmg/7.4.755. [DOI] [PubMed] [Google Scholar]
- Lea DE, Coulson CA. Distribution of numbers of mutants in bacterial population. Journal of Genetics. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang H, Veeraraghavan J, Bambara RA, Freudenreich CH. Saccharomyces cerevisiae flap endonuclease 1 uses flap equilibration to maintain triplet repeat stability. Mol Cell Biol. 2004;24:4049–4064. doi: 10.1128/MCB.24.9.4049-4064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf M, Ried K, Woollatt E, Dayan S, Eyre H, Finnis M, Hobson L, Nancarrow J, Venter D, Baker E, Richards RI. Chromosomal Fragile Site FRA16D and DNA Instability in Cancer. Cancer Res. 2000;60:1683–1689. [PubMed] [Google Scholar]
- McClellan JA, Boublikova P, Palecek E, Lilley DM. Superhelical torsion in cellular DNA responds directly to environmental and genetic factors. Proc Natl Acad Sci U S A. 1990;87:8373–8377. doi: 10.1073/pnas.87.21.8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin SM. DNA structures, repeat expansions and human hereditary disorders. Curr Opin Struct Biol. 2006;16:351–358. doi: 10.1016/j.sbi.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Mishmar D, Rahat A, Scherer SW, Nyakatura G, Hinzmann B, Kohwi Y, Mandel-Gutfroind Y, Lee JR, Drescher B, Sas DE, et al. Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc Natl Acad Sci U S A. 1998;95:8141–8146. doi: 10.1073/pnas.95.14.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe LV, Richards RI. Common chromosomal fragile sites and cancer: focus on FRA16D. Cancer Lett. 2006;232:37–47. doi: 10.1016/j.canlet.2005.07.041. [DOI] [PubMed] [Google Scholar]
- Palakodeti A, Han Y, Jiang Y, Le Beau MM. The role of late/slow replication of the FRA16D in common fragile site induction. Genes Chromosomes Cancer. 2004;39:71–76. doi: 10.1002/gcc.10290. [DOI] [PubMed] [Google Scholar]
- Pelletier R, Krasilnikova MM, Samadashwily GM, Lahue R, Mirkin SM. Replication and expansion of trinucleotide repeats in yeast. Mol Cell Biol. 2003;23:1349–1357. doi: 10.1128/MCB.23.4.1349-1357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards RI. Fragile and unstable chromosomes in cancer: causes and consequences. Trends in Genetics. 2001;17:339–345. doi: 10.1016/s0168-9525(01)02303-4. [DOI] [PubMed] [Google Scholar]
- Ried K, Finnis M, Hobson L, Mangelsdorf M, Dayan S, Nancarrow JK, Woollatt E, Kremmidiotis G, Gardner A, Venter D, et al. Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum Mol Genet. 2000;9:1651–1663. doi: 10.1093/hmg/9.11.1651. [DOI] [PubMed] [Google Scholar]
- Schwartz M, Zlotorynski E, Goldberg M, Ozeri E, Rahat A, le Sage C, Chen BP, Chen DJ, Agami R, Kerem B. Homologous recombination and nonhomologous end-joining repair pathways regulate fragile site stability. Genes Dev. 2005;19:2715–2726. doi: 10.1101/gad.340905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M, Zlotorynski E, Kerem B. The molecular basis of common and rare fragile sites. Cancer Lett. 2006;232:13–26. doi: 10.1016/j.canlet.2005.07.039. [DOI] [PubMed] [Google Scholar]
- Trenz K, Smith E, Smith S, Costanzo V. ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. Embo J. 2006;25:1764–1774. doi: 10.1038/sj.emboj.7601045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ND, Testa JR, Smith DI. Localization of three novel hybrid breakpoints and refinement of 18 marker assignments in the human 3cen-p21.1 region. Genomics. 1992;14:891–896. doi: 10.1016/s0888-7543(05)80110-7. [DOI] [PubMed] [Google Scholar]
- Zlotorynski E, Rahat A, Skaug J, Ben-Porat N, Ozeri E, Hershberg R, Levi A, Scherer SW, Margalit H, Kerem B. Molecular Basis for Expression of Common and Rare Fragile Sites. Mol Cell Biol. 2003;23:7143–7151. doi: 10.1128/MCB.23.20.7143-7151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.