Abstract
Under basal conditions, the interaction of the cytosolic protein Keap1 with the transcription factor Nrf2 results in a low level of expression of cytoprotective genes whose promoter region contains the antioxidant response element (ARE). Alkylation of one or more of the 27 cysteine sulfhydryl groups of human Keap1 is proposed to lead to Nrf2 nuclear accumulation, to upregulation of cytoprotective gene expression via the ARE, and to prevention of degenerative diseases, such as cancer. Therefore, identification of the most reactive of these cysteine residues towards specific electrophiles should help clarify this mechanism of cancer prevention, also known as chemoprevention. To address this issue, preliminary analyses of tryptic digests of Keap1 alkylated by the model electrophile 1-biotinamido-4-(4′-[maleimidoethyl-cyclohexane]-carboxamido) butane were carried out using LC-MS/MS with a cylindrical ion trap mass spectrometer and also using LC-MS/MS with a hybrid linear ion trap FT ICR mass spectrometer. Since the FT ICR instrument provided more complete peptide sequencing coverage and enabled the identification of more alkylated cysteine residues, only this instrument was used in subsequent studies of Keap1 alkylation by three electrophilic natural products that can up-regulate the ARE, xanthohumol, isoliquiritigenin and 10-shogaol. Among the various cysteine residues of Keap1, C151 was most reactive towards these three electrophiles. These in vitro results agree with evidence from in vivo experiments, and indicate that C151 is the most important site of alkylation on Keap1 by chemoprevention agents that function by activating the ARE through Nrf2.
Keywords: FT ICR MS, cysteine modification, Keap1, chemoprevention, xanthohumol, isoliquiritigenin, 10-shogaol
Introduction
Kelch-like ECH-associated protein 1 (Keap1) is a cytoplasmic protein that sequesters the transcription factor nuclear factor-E2-related factor 2 (Nrf2) in the cytosol under basal conditions [1]. Nrf2 is a member of the NF-E2 family of nuclear basic leucine zipper (bZIP) transcription factors. By binding to the 5′-upstream regulatory antioxidant response element (ARE) regions of cytoprotective genes, Nrf2 upregulates their transcription [2, 3], thus protecting cells against damage by reactive oxygen intermediates or other electrophilic species [4]. Keap1 functions as a bridge between Nrf2 and the Cullin3-based E3-ligase ubiquitination complex, promoting ubiquitination and subsequent proteasomal degradation of Nrf2 [5, 6, 7] thus preventing nuclear accumulation of Nrf2. Without the intervention of Keap1, levels of Nrf2 are elevated in the nucleus causing up-regulation of the ARE [8]. Alkylation of one or more of the 27 cysteine residues of Keap1 by reactive oxygen species and xenobiotic electrophiles, including chemopreventive agents, appears to be an important signaling mechanism for the regulation of the ARE via Nrf2 [3].
Keap1 has five distinct domains: the N-terminal domain (amino acids 1-60); the BTB domain (amino acids 61-178); a central linker domain (amino acids 179 to 321); the Kelch repeat domain (amino acids 322-608); and a C-terminal domain (amino acids 609-625). The BTB domain mediates the dimerization of Keap1 [9] and also binds the adaptor protein in Cul3-dependant ubiquitination systems [10]. The Kelch repeat domain binds to the Nrf2 directly [1]. Since Keap1 signalling is probably mediated by alkylation of one or more of its cysteine sulfhydryl groups [3, 11, 12], identification of the cysteine residues that are most reactive towards specific electrophiles should help clarify this mechanism of action of Keap1.
However, most of the work to date to identify reactive Keap1 cysteine residues has been carried out with model alkylating agents such as dexamethasone 21-mesylate [2], iodoacetyl-N-biotinyl hexylene diamine (BIA) and 1-biotinamido-4-(4′-[maleimidoethyl-cyclohexane]-carboxamido) butane (BMCC) [13] instead of biologically relevant ARE inducers that show promise as chemopreventive agents. Unlike ARE inducers that are Michael addition acceptors, alkylation by dexamethasone 21-mesylate and BIA is irreversible, which means that sample preparation would not lead to dissociation of such adducts. Since BMCC and BIA are biotinylated compounds, avidin-biotin affinity chromatography may be used to facilitate the isolation and identification of peptides alkylated by these electrophiles. However, few if any chemoprevention agents contain biotin tags to facilitate analysis. Therefore, a general analytical method for the characterization of alkylated Keap1 must minimize decomposition of adducts while facilitating the identification of alkylation sites without the benefit of affinity chromatography.
In this investigation, a method based on LC-MS/MS was developed that facilitated the detection of cysteine residues in human Keap1 modified by chemopreventive agents. Since it was anticipated that partial purification of the alkylated peptides from Keap1 might be necessary prior to LC-MS/MS analysis to detect all alkylated cysteine residues, preliminary analyses were carried out of Keap1 that had been alkylated by the model biotinylated electrophile BMCC using a cylindrical ion trap mass spectrometer (LCQ) or a linear ion trap-FI ICR mass spectrometer (LTQ-FT ICR). More BMCC alkylated cysteine residues in human Keap1 could be identified using the LTQ-FT ICR mass spectrometer without affinity isolation than when using either instrument with affinity purification. Therefore, this method was applied to the analysis of the reaction products between human Keap1 and each of three natural products containing Michael acceptors, xanthohumol, isoliquiritigenin and 10-shogaol (See structures in Scheme 1).
Scheme 1.
Structures of the natural product chemoprevention agents A) isoliquiritigenin [22]; B) xanthohumol [28]; and C) 10-shogaol [29]. The CD values represent the concentration of each compound required to double the intracellular quinone reductase activity in Hepa 1c1c7 murine hepatoma cells [22] and indicate the relative activities of these compounds as chemopreventive reagents.
Isolated from the hop plant, Humulus lupulus L. (Cannabaceae), xanthohumol has been reported to be a cancer chemopreventive agent with activity both in vitro and in vivo [14, 15, 16, 17]. Isoliquiritigenin is a constituent of the tonka bean Dipteryx odorata (Aubl.) Willd, and is also present in licorice (Glycyrrhiza uralensis) [18]. Isoliquiritigenin has been reported to have chemoprevention activity in vitro as well as several animal models [19, 20, 21, 22]. 10-Shogaol is a constituent of the rhizome of Zingiber officinale (ginger) [23]. Although not attributed specifically to 10-shogaol, ginger has been reported to have antioxidant and anticarcinogenic properties by several groups [24, 25, 26, 27]. All three of these natural products contain electrophilic unsaturated ketones (Scheme 1). As an indication of their chemopreventive activity through the induction of ARE, the induction of the phase 2 enzyme quinone reductase in Hepa 1c1c7 murine hepatoma cells by xanthohumol, isoliquiritigenin and 10-shogaol has been determined to be 7 ± 0.7, 1.8 ± 0.44 and 14.1 ± 0.7 μM respectively [22, 28, 29]. In this investigation, the relative reactivities of each of these compounds towards specific cysteine residues of human Keap1 were determined using LC-MS/MS, and the most reactive amino acid residue was identified as C151 for all three compounds.
Experimental
Materials and reagents
Recombinant human Keap1 protein containing a histidine tag was expressed in and purified from Escherichia coli. The details of the cloning, expression, and purification of human Keap1 are described elsewhere [30]. BMCC was purchased from Pierce (Rockford, IL), and trypsin was purchased from Promega (Madison, WI). An ICAT™ kit for protein labeling was purchased from Applied Biosystems (Foster City, CA). Deionized water was prepared using a Milli-Q purification system (Millipore, S.A. France). Acetonitrile (Optima) was purchased from Fisher Scientific (Hanover Park, IL). Isoliquiritigenin, dimethyl sulfoxide, tris(carboxylethyl)phosphine, D,L-dithiothreitol (DTT), and iodoacetamide were purchased from Sigma-Aldrich (St. Louis, MO, and Milwaukee WI). 10-Shogaol was isolated and provided by Dr. Yi Tao of the University of Illinois College of Pharmacy, and xanthohumol was isolated and provided by Dr. Luke R. Chadwick and Dr. Guido F. Pauli of the same institution.
Sample preparation
Alkylation of Keap1 by isoliquiritigenin, xanthohumol, 10-shogaol, or BMCC (as a positive control) was carried out at different ratios as follows (using BMCC as an example). A stock solution of BMCC was prepared at a concentration of 10 μM in dimethyl sulfoxide. Human Keap1 (10 μM) was incubated with BMCC at molar ratios from 1:0.025 to 1:2 ([Keap1]/[BMCC]) at room temperature for 2 h in 100 μL 25 mM Tris buffer (pH 8.0). Next, DTT was added to this reaction mixture at a concentration of 1 mM. After incubation for 30 min, excess iodoacetamide was added to block the remaining free cysteines in Keap1. Subsequently, 5 mM DTT was added to quench unreacted iodoacetamide. Finally, 0.5 μg trypsin was added to each Keap1 solution followed by incubation at 37°C for 3 h (BMCC) or 1.5 h (all other electrophiles). The tryptic peptides were analyzed using LC-MS/MS. Alternatively, BMCC modified peptides were purified from the tryptic digest of Keap1 using an avidin affinity column as described by the manufacturer of the ICAT kit before analysis by LC-MS/MS.
LC-MS/MS analyses using a cylindrical ion trap mass spectrometer
Peptides were analyzed initially using a Finnigan LCQDeca (Thermo, San Jose, CA) ion trap mass spectrometer equipped with a Surveyor HPLC system, and a Vydac (Hesperia, CA) 218MS LC-MS C18 reversed phase HPLC column (5 μM, 2.1 × 150 mm, 300 Å) and a 218TP (5 μM, 2.1 mm) guard column. The solvent system consisted of a linear gradient from 5 – 45% solvent B over 60 min and then 45 – 80% solvent B over 15 min (solvent A: 95:4.9:0.1; and solvent B: 4.9:95:0.1, water/acetonitrile/formic acid, v/v/v) at a flow rate of 200 μL/min. The LCQ mass spectrometer was operated in a data-dependent MS/MS mode in which the most abundant peptide ion in each mass spectrum was selected for collision-induced dissociation using a normalized collision energy of 35%. The LC-MS/MS data were processed using Thermo BioWorks™ 3.1 and TurboSEQUEST. The sites of Keap1 alkylation were identified based on both automated data processing and manual inspection of the tandem mass spectra.
LC-MS/MS analyses using a linear ion trap-FT ICR hybrid mass spectrometer
For confirmation of the data obtained using the cylindrical ion trap, LC-MS/MS analysis of the alkylated Keap1 peptides was also carried out using a high resolution Finnigan LTQ-FT ICR mass spectrometer equipped with a Dionex (Auburn, CA) microcapillary HPLC system. The same HPLC separation conditions were used as described above except that a 35 min linear gradient was used from the 5–45% solvent B. The LTQ-FT ICR mass spectrometer was operated in a data-dependent MS/MS mode in which the 5 most abundant peptide ions in each mass spectrum were selected for collision-induced dissociation using a normalized collision energy of 35%.
Results and Discussion
Cysteines in Keap1 modified by BMCC
Because Keap1 has a molecular mass exceeding 70,000 Da, tryptic digestion formed in excess of 100 peptides (calculated using one missing cleavage site). To detect all the modified peptides, it was anticipated that partial purification of the modified peptides would be necessary such as ion exchange fractionation or affinity extraction prior to LC-MS/MS analysis. To determine whether affinity purification step was necessary, a preliminary study was carried out using Keap1 that had been alkylated by the model electrophilic biotinylated molecule BMCC using a cylindrical ion trap mass spectrometer (LCQ). As shown in Table 1, avidin purification usually increased the number of modified peptides detected, though not to a great extent.
Table 1.
Alkylation of cysteine residues in human Kelch-like ECH-associated protein 1 (Keap1) by 1-biotinamido-4-(4′-[maleimidoethyl-cyclohexane]-carboxamido) butane (BMCC) as a function of BMCC concentration. BMCC was incubated with Keap1 at different molar ratios, and at each molar ratio, the tryptic peptides of Keap1 were analyzed using two instruments: a cylindrical ion trap mass spectrometer and a linear ion trap-FT ICR mass spectrometer.
| [BMCC]/[Keap1] | Avidin Purification | Modified cysteines detected using the cylindrical ion trap MS | Modified cysteines detected using the linear ion trap-FT ICR MS |
|---|---|---|---|
| 0.025 | No | Nonea | C151, C434b |
| 0.025 | Yes | None | None |
| 0.05 | No | None | C151, C434 |
| 0.05 | Yes | None | None |
| 0.1 | No | None | C151, C434, C288, C319, C613 |
| 0.1 | Yes | None | None |
| 0.5 | No | C151 | C151, C434, C288, C319, C613, C226, C257, C297 |
| 0.5 | Yes | None | C434, C288, C319, C297. |
| 1 | No | C151, C319, C273, C257, C297 | C151, C434, C288, C319, C613, C226, C257, C297, C77, C273 |
| 1 | Yes | C151, C319, C273, C257, C297, C613, C226 | C151, C434, C288, C319, C613, C226, C257, C297, C77, C273 |
| 2 | No | C151, C319, C273, C257, C297, C613, C226, C434 | C151, C434, C288, C319, C613, C226, C257, C297, C77, C273, C23, C38 |
| 2 | Yes | C151, C319, C273, C257, C297, C613, C226, C434, C622, C288, C241, C23 | C151, C434, C288, C319, C613, C226, C257, C297, C77, C273, C23, C38 |
“None” indicates that no peptides containing BMCC modified cysteine residues were detected.
Peptides detected containing BMCC modified cysteine. The order in which these modified cysteines are listed corresponds to their relative abundances.
Upon acquisition of a linear ion trap FT ICR mass spectrometer (LTQ-FT ICR MS), the study was updated using this new instrument for LC-MS/MS analysis. Due to its faster scan speed and greater sensitivity, more biotinylated peptides were identified using the LTQ-FT ICR than the LCQ mass spectrometer, especially when the ratio of BMCC to protein was low (See Table 1). Routinely, 90% of the protein sequence was covered by MS/MS analysis using the LTQ-FT ICR mass spectrometer (compared to only 75 to 80% of sequence coverage using the LCQ mass spectrometer), and all the cysteine-containing tryptic peptides were detected. Comparing the alkylation sites found by using the LCQ and LTQ-FT mass spectrometers at the same molar ratio of BMCC and Keap1, more modified sites were detected using the LTQ-FT mass spectrometer (Table 1). Interestingly, at low ratios, avidin purification actually reduced the number of detected peptides. This might be due to the high affinity of the biotin-avidin complex (Kd ~10−15 M−1) [31], which perhaps retained some of the biotinylated peptides on the avidin column, and caused some BMCC modified peptides to be lost during the affinity purification. A more probable possible cause of the loss of BMCC modified peptides is dissociation of the BMCC-cysteine adducts during affinity purification. Since BMCC is a Michael addition acceptor, it can modify cysteine residues reversibly. Therefore, during avidin-biotin affinity purification, some of the adducts probably dissociated before they could be detected.
FT ICR mass spectrometry greatly facilitated the identification of alkylated cysteine residues in human Keap1, in particular, those that could not be purified by affinity means. Therefore, this FT ICR mass spectrometer-based method was used without affinity column purification in all subsequent experiments for the analysis of the reactivities of Keap1 cysteines towards ARE inducers.
Modification sites of human Keap1 by electrophiles
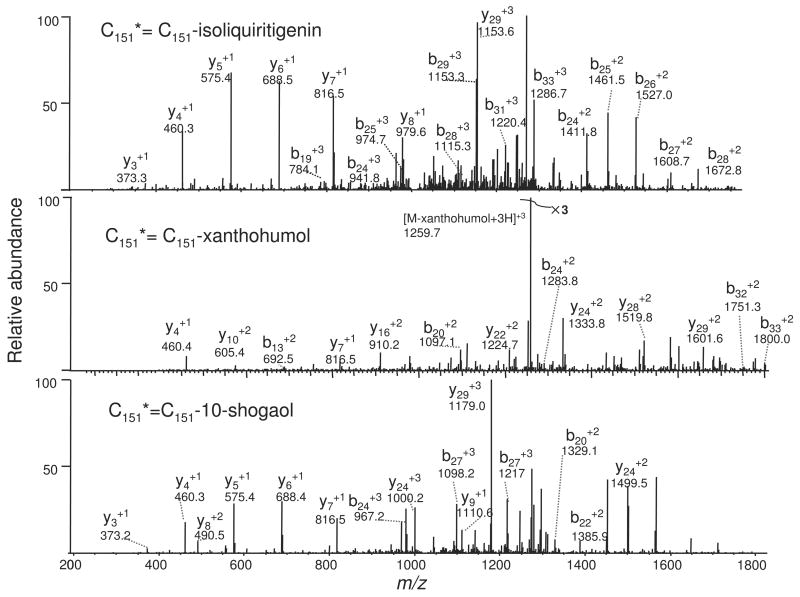
Since FT ICR LC-MS/MS facilitated the identification of sites of cysteine alkylation by electrophiles in human Keap1 without the requirement of affinity column enrichment of alkylated peptides, this instrument was used for the detection of the alkylation sites in Keap1 after reaction with electrophiles lacking affinity tags such as biotin. Specifically, this method was used to determine the modification pattern of Keap1 after alkylation by ARE inducers containing a Michael addition moiety. Three different electrophilic natural products, isoliquiritigenin, xanthohumol and 10-shogaol (Scheme 1) were incubated with Keap1. Incubations with a range of concentrations of these electrophiles were used to determine through a titration-like process which Keap1 cysteine residues became alkylated first. Trypsin digestion followed by peptide mapping and sequencing of the alkylated Keap1 protein were carried out to determine the sites of alkylation. As expected, only cysteine residues were alkylated, and these residues are shown in Table 2. The three most readily modified cysteines of human Keap1 by xanthohumol were identified as C151, C319 and C613. The two most readily modified cysteines of human Keap1 by isoliquiritigenin were identified as C151 and C226. The three most readily modified cysteines of human Keap1 by 10-shogaol were C151, C257 and C368. The tandem mass spectra of the peptide ions containing C151 residues that had been alkylated by xanthohumol, isoliquiritigenin or 10-shogaol are shown in Figure 1.
Table 2.
Cysteine residues in Keap1 modified by chemopreventive agents as indicated by LC-MS/MS analysis.
| Domain | Cysteine | [Xanthohumol]/[Keap1]
|
[Isoliquiritigenin]/[Keap1]
|
[10-Shogaol]/[Keap1]
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 10 | 20 | 2 | 5 | 10 | 2 | 5 | 10 | ||
| N-terminal | C13 | 1,2 | ||||||||
| C14 | 1,2 | |||||||||
| C23 | 1,2 | 1,2,3 | 2,3 | |||||||
| C38 | 1,2 | 1 | 2,3 | |||||||
|
| ||||||||||
| BTB | C77 | 3 | 3 | 1,2,3 | 1,2,3 | |||||
| C151 | 1,2,3 | 1,2,3 | 1,2,3 | 1,2,3 | 1,2,3 | 1,2,3 | 1,2,3 | 1,2,3 | 1,2,3 | |
| C171 | ||||||||||
|
| ||||||||||
| Central linker | C196 | 1,2 | 1,2,3 | |||||||
| C226 | 1,2 | 1,2,3 | 1,2,3 | 1,2,3 | ||||||
| C241 | 1,2,3 | 1,2,3 | ||||||||
| C249 | 1 | 1,2,3 | 1,2,3 | |||||||
| C257 | 1,2,3 | 1,2,3 | 1,2,3 | |||||||
| C273 | 1,3 | |||||||||
| C288 | ||||||||||
| C297 | ||||||||||
| C319 | 1,2,3 | 1,2,3 | 1,2 | 3 | 1,2,3 | 1,2,3 | 1,2,3 | 1,2,3 | ||
|
| ||||||||||
| Kelch | C368 | 1,3 | 1,2,3 | 1,2,3 | 1,2,3 | |||||
| C395 | 1,2 | 1,2,3 | ||||||||
| C406 | ||||||||||
| C434 | 1,2 | 1,2,3 | 1,3 | 1,2,3 | 1,2,3 | |||||
| C489 | 1,2,3 | 2 | 1,2,3 | 1,2,3 | ||||||
| C513 | 3 | 1,2 | 1,2,3 | |||||||
| C518 | 1,2 | 1,2,3 | ||||||||
| C583 | 2 | 1,2,3 | 1,2,3 | |||||||
|
| ||||||||||
| C-terminal | C613 | 1,2,3 | 1,2,3 | 1,2,3 | 3 | 1,2,3 | 1,2,3 | 1,2,3 | ||
| C622 | 1,2,3 | 1,2,3 | ||||||||
| C624 | 1,2,3 | 1,2,3 | ||||||||
These data represent the results of LC-MS/MS analyses of three identical experiments. The numbers indicate the experiment numbers (1, 2 and 3) in which specific modified cysteines were detected.
Figure 1.
MS/MS spectra of alkylated Keap1 C151 peptides of the sequence LIEFAYTASISMGEKC151*VLHVMNGAVMYQIDSVVR. Keap1 was alkylated by the electrophilic natural products xanthohumol, isoliquiritigenin or 10-shogaol and then digested with trypsin. Cys151* represents the site of modification by each electrophilic natural product. All of the peptide fragment ions y19 to y33 and b16 to b33 contained the modification.
Prior to the discovery of Keap1 protein, Talalay et al. [32] had proposed that there must be a protein with highly reactive cysteine thiols that acts as a sensor for phase 2 inducers. Since the isolation of Keap1 from a yeast system using a Gal4-Neh2 fusion protein [33], great interest has focused on the reactivity of Keap1 cysteine residues. Dinkova-Kostova et al. [2] studied the reaction of murine Keap1 with dexamethasone mesylate using mass spectrometry and suggested that C257, C273, C288 and C297 were the most reactive sites among the 25 murine Keap1 cysteine residues. In their studies of murine Keap1, Dinkova-Kostova et al. [2] used MALDI-TOF to identify modified peptides in a tryptic digest, but LC-MS/MS was used to confirm the sequence and to determine the site of modification in only two of the four modified peptides. Our previous study [30] in which human Keap1 modifications were mapped hindicated that C151, C288 and C297 are the most reactive residues towards iodoacetyl-N-biotinyl hexylene diamine. A similar study was reported by Hong et al. [13], but their data suggested that C169, C241 and C288 but not C151 were reactive towards human Keap1. Subsequently, we found that a limitation of the sample preparation procedure used by Hong et al. [13] had resulted in their inability to detect the modified C151 peptide [36]. Specifically, Hong et al. [13] used an ultracentrifugation device for buffer exchange, which resulted in the binding and loss of >99% of the protein due to adsorption and possibly changed the conformation of Keap1 so that a different modification pattern was detected [36].
We have developed an LC-MS/MS method to characterize the reactivity of human Keap1 towards Michael addition reaction acceptors from natural products that upregulate the ARE. Furthermore, this approach facilitated the identification of the most reactive amino acid residues in Keap1. Although the Keap1 alkylation pattern was unique for each electrophile, only cysteine residues were alkylated, and C151 was always detected among the top 3 most reactive cysteines. At a ratio of electrophile to Keap1 of only 2:1, which was the smallest ratio tested, 10-shogal and isoliquiritigen but not xanthohumol alkylated C151 to a detectable level. As the ratio of electrophile to Keap1 in the incubation was increased, cysteine modifications by isoliquiritigen and 10-shogal were detected on all 5 protein domains. Xanthohumol was the least reactive of the three electrophiles, there were no modification sites detected at a ratio of 2 to 1, but at a ratio 5 to 1, C151 (located in BTB domain), and C319 (located in central linker domain) and C613 (located in Kelch repeat domain) were modified.
The differences among adduct patterns of these three electrophiles may be due to their unique structures and reactivities. Regardless, these results indicate C151 is extremely reactive towards these three chemoprevention agents. Located in the BTB domain of Keap1, site-directed mutagenesis studies have indicated that C151 is essential for inhibition of Keap1-dependant ubiquitination and degradation of Nrf2 in response to all tested agents, including tBHQ and sulforaphane [3], NEPP11, an endogenous neurite outgrowth-promoting prostaglandin [34], and ebselen, a seleno-organic drug [35]. Taken together, C151 plays a critical role in the Keap1-Nrf2 signaling system, and our results support the view that C151 is a target for Nrf2 activation by Michael addition reaction acceptors that function as ARE inducers.
Conclusions
The use of FT ICR LC-MS-MS facilitated the identification of sites of cysteine alkylation by electrophiles in human Keap1 digested with trypsin without the need for affinity column enrichment of alkylated peptides. The reaction of Keap1 with the Michael addition reaction acceptors xanthohumol, isoliquiritigenin and 10-shogaol, which are natural products that up-regulate the ARE, was investigated using this method. All three compounds alkylated Keap1 at specific cysteine residues, and the most reactive site of Keap1 was C151. This information suggests that natural product chemoprevention agents that are Michael addition reaction acceptors up-regulate the ARE as a result of alkylation of Keap1 at C151.
Acknowledgments
This work was support by grant number P01 CA48112 from the National Cancer Institute and grant P50 AT00155 provided to the UIC/NIH Center for Botanical Dietary Supplements Research by the Office of Dietary Supplements, the Office for Research on Women’s Health, and the National Center for Complementary and Alternative Medicine. The authors thank the Chicago Biomedical Consortium for access to the LTQ-FT ICR mass spectrometer which was purchased with a grant from The Searle Funds at The Chicago Community Trust.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 Represses Nuclear Activation of Antioxidant Responsive Elements by Nrf2 through Binding to the Amino-Terminal Neh2 Domain. Genes Dev. 1999;1:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct Evidence that Sulfhydryl Groups of Keap1 are the Sensors Regulating Induction of Phase 2 Enzymes that Protect Against Carcinogens and Oxidants. Proc Natl Acad Sci U S A. 2002;18:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang DD, Hannink M. Distinct Cysteine Residues in Keap1 are Required for Keap1-Dependent Ubiquitination of Nrf2 and for Stabilization of Nrf2 by Chemopreventive Agents and Oxidative Stress. Mol Cell Biol. 2003;22:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Talalay P, Fahey JW, Holtzclaw WD, Prestera T, Zhang Y. Chemoprotection Against Cancer by Phase 2 Enzyme Induction. Toxicol Lett. 1995:173–179. doi: 10.1016/0378-4274(95)03553-2. [DOI] [PubMed] [Google Scholar]
- 5.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB Protein is an Adaptor that Bridges Nrf2 to a Cul3-Based E3 Ligase: Oxidative Stress Sensing by a Cul3-Keap1 Ligase. Mol Cell Biol. 2004;19:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furukawa M, Xiong Y. BTB Protein Keap1 Targets Antioxidant Transcription Factor Nrf2 for Ubiquitination by the Cullin 3-Roc1 Ligase. Mol Cell Biol. 2005;1:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a Redox-Regulated Substrate Adaptor Protein for a Cul3-Dependent Ubiquitin Ligase Complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itoh K, Wakabayashi N, Katoh Y, Ishii T, O’Connor T, Yamamoto M. Keap1 Regulates both Cytoplasmic-Nuclear Shuttling and Degradation of Nrf2 in Response to Electrophiles. Genes Cells. 2003;4:379–391. doi: 10.1046/j.1365-2443.2003.00640.x. [DOI] [PubMed] [Google Scholar]
- 9.Zipper LM, Mulcahy RT. The Keap1 BTB/POZ Dimerization Function is Required to Sequester Nrf2 in Cytoplasm. J Biol Chem. 2002;39:36544–36552. doi: 10.1074/jbc.M206530200. [DOI] [PubMed] [Google Scholar]
- 10.Dinkova-Kostova AT, Holtzclaw WD, Kensler TW. The Role of Keap1 in Cellular Protective Responses. Chem Res Toxicol. 2005;12:1779–1791. doi: 10.1021/tx050217c. [DOI] [PubMed] [Google Scholar]
- 11.Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, Kensler TW, Talalay P. Protection Against Electrophile and Oxidant Stress by Induction of the Phase 2 Response: Fate of Cysteines of the Keap1 Sensor Modified by Inducers. Proc Natl Acad Sci U S A. 2004;7:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong F, Freeman ML, Liebler DC. Identification of Sensor Cysteines in Human Keap1 Modified by the Cancer Chemopreventive Agent Sulforaphane. Chem Res Toxicol. 2005;12:1917–1926. doi: 10.1021/tx0502138. [DOI] [PubMed] [Google Scholar]
- 13.Hong F, Sekhar KR, Freeman ML, Liebler DC. Specific Patterns of Electrophile Adduction Trigger Keap1 Ubiquitination and Nrf2 Activation. J Biol Chem. 2005;36:31768–31775. doi: 10.1074/jbc.M503346200. [DOI] [PubMed] [Google Scholar]
- 14.Gerhauser C. Broad Spectrum Anti-Infective Potential of Xanthohumol from Hop (Humulus Lupulus L.) in Comparison with Activities of Other Hop Constituents and Xanthohumol Metabolites. Mol Nutr Food Res. 2005;9:827–831. doi: 10.1002/mnfr.200500091. [DOI] [PubMed] [Google Scholar]
- 15.Gerhauser C. Beer Constituents as Potential Cancer Chemopreventive Agents. Eur J Cancer. 2005;13:1941–1954. doi: 10.1016/j.ejca.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 16.Stevens JF, Page JE. Xanthohumol and Related Prenylflavonoids from Hops and Beer: To Your Good Health! Phytochemistry. 2004;10:1317–1330. doi: 10.1016/j.phytochem.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 17.Albini A, Dell’Eva R, Vene R, Ferrari N, Buhler DR, Noonan DM, Fassina G. Mechanisms of the Antiangiogenic Activity by the Hop Flavonoid Xanthohumol: NF-kappaB and Akt as Targets. FASEB J. 2006;3:527–529. doi: 10.1096/fj.05-5128fje. [DOI] [PubMed] [Google Scholar]
- 18.Kim DC, Choi SY, Kim SH, Yun BS, Yoo ID, Reddy NR, Yoon HS, Kim KT. Isoliquiritigenin Selectively Inhibits H(2) Histamine Receptor Signaling. Mol Pharmacol. 2006;2:493–500. doi: 10.1124/mol.106.023226. [DOI] [PubMed] [Google Scholar]
- 19.Jang DS, Park EJ, Kang YH, Hawthorne ME, Vigo JS, Graham JG, Cabieses F, Fong HH, Mehta RG, Pezzuto JM, Kinghorn AD. Potential Cancer Chemopreventive Flavonoids from the Stems of Tephrosia Toxicaria. J Nat Prod. 2003;9:1166–1170. doi: 10.1021/np0302100. [DOI] [PubMed] [Google Scholar]
- 20.Baba M, Asano R, Takigami I, Takahashi T, Ohmura M, Okada Y, Sugimoto H, Arika T, Nishino H, Okuyama T. Studies on Cancer Chemoprevention by Traditional Folk Medicines XXV. Inhibitory Effect of Isoliquiritigenin on Azoxymethane-Induced Murine Colon Aberrant Crypt Focus Formation and Carcinogenesis. Biol Pharm Bull. 2002;2:247–250. doi: 10.1248/bpb.25.247. [DOI] [PubMed] [Google Scholar]
- 21.Yamazaki S, Morita T, Endo H, Hamamoto T, Baba M, Joichi Y, Kaneko S, Okada Y, Okuyama T, Nishino H, Tokue A. Isoliquiritigenin Suppresses Pulmonary Metastasis of Mouse Renal Cell Carcinoma. Cancer Lett. 2002;1:23–30. doi: 10.1016/s0304-3835(02)00113-1. [DOI] [PubMed] [Google Scholar]
- 22.Cuendet M, Oteham CP, Moon RC, Pezzuto JM. Quinone Reductase Induction as a Biomarker for Cancer Chemoprevention. J Nat Prod. 2006;3:460–463. doi: 10.1021/np050362q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwasaki Y, Morita A, Iwasawa T, Kobata K, Sekiwa Y, Morimitsu Y, Kubota K, Watanabe T. A Nonpungent Component of Steamed Ginger--[10]-Shogaol--Increases Adrenaline Secretion Via the Activation of TRPV1. Nutr Neurosci. 2006;3–4:169–178. doi: 10.1080/110284150600955164. [DOI] [PubMed] [Google Scholar]
- 24.Burton A. Chemoprevention: Eat Ginger, Rub on Pomegranate. Lancet Oncol. 2003;12:715. doi: 10.1016/s1470-2045(03)01295-6. [DOI] [PubMed] [Google Scholar]
- 25.Ihlaseh SM, de Oliveira ML, Teran E, de Camargo JL, Barbisan LF. Chemopreventive Property of Dietary Ginger in Rat Urinary Bladder Chemical Carcinogenesis. World J Urol. 2006;5:591–596. doi: 10.1007/s00345-006-0108-9. [DOI] [PubMed] [Google Scholar]
- 26.Manju V, Nalini N. Chemopreventive Efficacy of Ginger, a Naturally Occurring Anticarcinogen during the Initiation, Post-Initiation Stages of 1,2 Dimethylhydrazine-Induced Colon Cancer. Clin Chim Acta. 2005;1–2:60–67. doi: 10.1016/j.cccn.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 27.Park EJ, Pezzuto JM. Botanicals in Cancer Chemoprevention. Cancer Metastasis Rev. 2002;3–4:231–255. doi: 10.1023/a:1021254725842. [DOI] [PubMed] [Google Scholar]
- 28.Dietz BM, Kang YH, Liu G, Eggler AL, Yao P, Chadwick LR, Pauli GF, Farnsworth NR, Mesecar AD, van Breemen RB, Bolton JL. Xanthohumol Isolated from Humulus Lupulus Inhibits Menadione-Induced DNA Damage through Induction of Quinone Reductase. Chem Res Toxicol. 2005;8:1296–1305. doi: 10.1021/tx050058x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao Y. Ph D dissertation. University of Illinois; Chicago: 2007. Characterization of Cyclooxygenase-2 Inhibitors as Anti-Inflammatory Agents from Ginger Dietary Supplements and in vitro Metabolism Studies of Gingerol-Related Compounds; p. 137. [Google Scholar]
- 30.Eggler AL, Liu G, Pezzuto JM, van Breemen RB, Mesecar AD. Modifying Specific Cysteines of the Electrophile-Sensing Human Keap1 Protein is Insufficient to Disrupt Binding to the Nrf2 Domain Neh2. Proc Natl Acad Sci U S A. 2005;29:10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rybak JN, Scheurer SB, Neri D, Elia G. Purification of Biotinylated Proteins on Streptavidin Resin: A Protocol for Quantitative Elution. Proteomics. 2004;8:2296–2299. doi: 10.1002/pmic.200300780. [DOI] [PubMed] [Google Scholar]
- 32.Talalay P, De Long MJ, Prochaska HJ. Identification of a Common Chemical Signal Regulating the Induction of Enzymes that Protect Against Chemical Carcinogenesis. Proc Natl Acad Sci U S A. 1988;21:8261–8265. doi: 10.1073/pnas.85.21.8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xue F, Cooley L. Kelch Encodes a Component of Intercellular Bridges in Drosophila Egg Chambers. Cell. 1993;5:681–693. doi: 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]
- 34.Santos RL, Hassan MJ, Sikandar S, Lee K, Ali G, Martin PE, Jr, Wambangco MA, Ahmad W, Leal SM. DFNB68, a Novel Autosomal Recessive Non-Syndromic Hearing Impairment Locus at Chromosomal Region 19p13.2. Hum Genet. 2006;1:85–92. doi: 10.1007/s00439-006-0188-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakurai T, Kanayama M, Shibata T, Itoh K, Kobayashi A, Yamamoto M, Uchida K. Ebselen a Seleno-Organic Antioxidant as an Electrophile. Chem Res Toxicol. 2006;9:1196–1204. doi: 10.1021/tx0601105. [DOI] [PubMed] [Google Scholar]
- 36.Eggler A, Luo Y, van Breemen RB, Mesecar AD. Identification of the Highly Reactive Cysteine 151 in the Chemopreventive Agent-Sensor Keap1 Protein is Method-Dependent. Chem Res Toxicol. 2007 doi: 10.1021/tx700217c. in press. [DOI] [PubMed] [Google Scholar]