Synopsis
This chapter reviews the molecular biology of the hepatitis B virus (HBV) in an effort to explain its natural history from a molecular perspective. The life cycle of the virus, with special attention to virus replication, polypeptide production and morphogenesis, is described. The way in which these steps may influence the natural history of viral pathogenesis, as well as the effectiveness of interventions, receives special consideration.
Introduction
HBV is a small, enveloped, hepatotropic virus with a partly double-stranded, relaxed circular (rc) DNA genome [1, 2]. It is the prototype member of the hepadnaviridae family. Although the animal hepadnaviruses, including woodchuck hepatitis virus (WHV), duck hepatitis B virus (HBV) as well as others, do not establish infections in people, they provide model systems for the study of viral replication, pathogenesis and evaluation of antiviral drugs against HBV [3, 4].
Natural infection with HBV can be transient with resolution or result in chronic infection [2, 5]. Transient infection may be characterized by acute hepatitis and, in rare cases, fatal fulminant hepatitis [5]. Chronic infection is a major public health burden affecting an estimated 350 – 400 million individuals worldwide and carries a high risk for the development of chronic active hepatitis, cirrhosis and primary hepatocellular carcinoma (HCC) [6]. Since the time between infection and clinical illness in chronic infections is usually decades, there is a significant opportunity for therapeutic intervention.
Hepatitis B virus and its infection in vivo and in vitro
Virion components
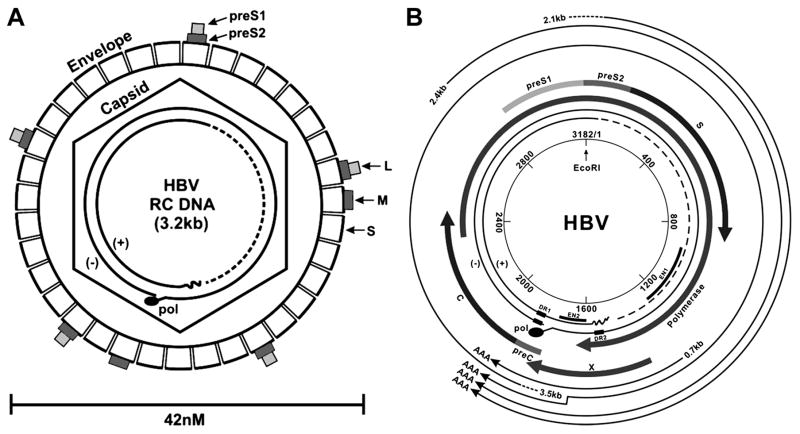
As Fig. 1 illustrates, the infectious HBV virion is approximately 42 nM in diameter and comprised of ~3.2 kb partially double-stranded relaxed, circular DNA (rcDNA) genome within a nucleocapsid (core) that is surrounded by a lipid bilayer studded with complexes of viral glycoproteins [1]. The nucleocapsid is a 27 nm diameter icosahedron assembled from 240 viral capsid proteins and packages a single copy of viral genomic DNA and DNA polymerase that is covalently linked to the 5’ end of minus strand DNA [7, 8]. There is evidence that cellular proteins, including chaperones and protein kinases, are also packaged inside of nucleocapsids [9]. The nucleocapsid is enveloped during its budding from the endoplasmic reticulum and the envelope membrane contains three viral glycoproteins, called large (L, LHBs), medium (M, MHBs) and small (S, SHBs) surface antigens that are translated from the different in-frame starting codons within the same open reading frame [1].
Figure 1. The HBV virion and genomic structural features.
(A) Illustration of the HBV virion (1A), showing envelope, S, M, L, with their S, preS2 and preS1 domains in sub-boxes of size proportional to their polypeptide length. The HBV genome, contained within the capsid, is shown as a partly double stranded, incomplete circle, with the polymerase protein covalently attached to the “minus” strand. (B) The circular map of the HBV genome is shown, with nucleotides numbered from the single EcoR1 site (by convention) and transcripts and their polypeptide products indicated.
Infected cells usually secrete 100 to 1000 times as many empty polymers of envelope proteins with spherical or filamentous shapes made mostly of SHBs (and less MHBs) as they do infectious virions [2]. The significance of the empty SHBs is unclear, and has been suspected to play a role in immunoevasion [10, 11]. In any event, the presence of SHBs in the circulation over a period of more than 6 months is usually accepted as evidence of chronic infection, and it is SHBs that most commercial assays for virus envelope protein are detecting.
Mode of transmission and host range
HBV is transmitted from person to person, via “sex, blood and needles” (Hepatitis B Foundation web site, www.hepb.org). Most chronic carriers of the virus in the world acquired their infection neonatally from their infected mothers. There have been reports of transmission via blood consuming and sequestering arthropods, but its public health significance has not been established. Taken together, most transmission can be accounted for by person to person contact through parenteral routes; however, the source of as many as one-third of the transmissions remains unclear. There are no natural animal intermediates or vectors, although replication is supported in chimpanzees and possibly tupaia [12, 13].
Infection in vivo is extremely efficient, but infection of cultured hepatocytes has been difficult to demonstrate
HBV infection appears to be highly efficient in vivo. As mentioned above, people can be infected with contaminated needles that usually represent an inoculation of less than 100 infectious viral particles. Furthermore, inoculation of chimpanzees or ducks with a single infectious HBV or DHBV particle, respectively, is sufficient to establish an infection [14, 15]. In contrast, infection of tissue culture lines of transformed liver cancer cells as well as primary human and chimpanzee hepatocytes, maintained as monolayers in culture dishes, with HBV is extremely inefficient and particularly difficult [16]. The reasons for this striking discrepancy in vivo and in vitro are not known and especially puzzling since HBV DNA introduced into cultured cells can result in production of infectious virus [17, 18]. One possibility is that hepatocytes and malignantly transformed cells, explanted from the organ and grown in tissue culture, lose properties essential to the early steps in the virus life cycle such as receptors and or co-receptors. Perhaps hepatocyte polarity is important for viral entry and this is lost in culture.
However, HBV replication is supported by hepatoblastoma and hepatoma cell lines. For example, transfection of plasmids containing infection competent HBV genomes into human hepatoma Huh7 and hepatoblastoma HepG2 cultures results in production and secretion of enveloped virions [17, 19]. These systems allow for the study of HBV RNA transcription, DNA synthesis, and the assembly and secretion of viral particles which occur following transfection, but the early events of virus infection of cells such as receptor binding, entry, capsid disassembly and the first-round cccDNA formation can not be studied. The recently reported tissue culture infection system described by Gripon and colleagues of a highly differentiated hepatocyte-derived cell line (HepaRG) that can be infected with serum derived HBV may provide a new means to address the early steps in the virus life cycle [20].
HBV Life Cycle
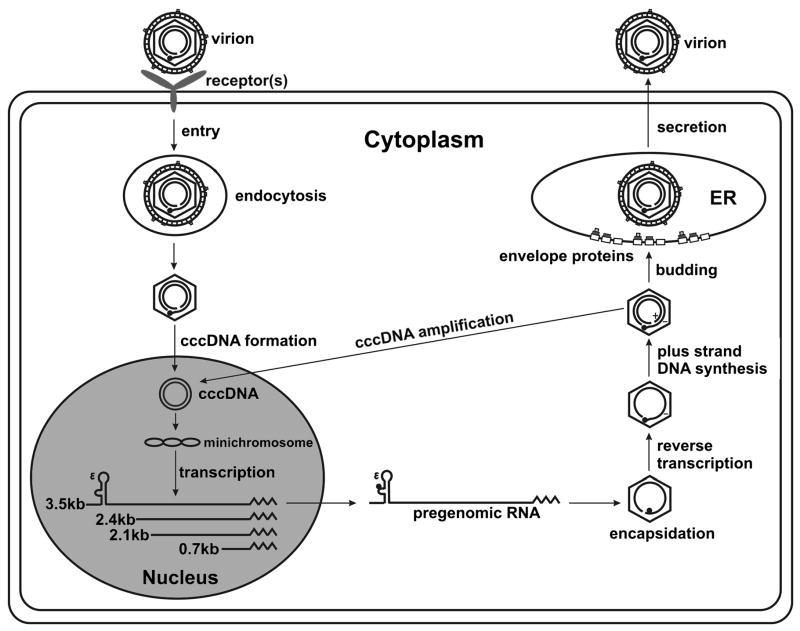
HBV has remarkable properties that make it fascinating as a research model as well as provide clues about its mechanisms of pathogenesis. For example, although its genome, as isolated from virion, is 3.2 kb of mostly double-stranded DNA (Fig. 1A), its life cycle is dependent upon reverse transcription of a longer than full length RNA copy of its genome called pre-genomic (pg) RNA [1]. Thus, HBV is a retrovirus with a DNA genome. HBV is also the major etiological agent of hepatocellular carcinoma, despite containing no known oncogenes. A simplified outline of the HBV replication cycle is shown in Figure 2.
Figure 2. A schematic outline of the HBV replication cycle.
Major steps in the molecular biology of the HBV replication life cycle are shown, from attachment to translocation of the virion DNA to the nucleus, to conversion of entering viral DNA into cccDNA, followed by transcription of the cccDNA into the viral RNA gene products. Encapsidations in the cytoplasm, virion morphogenesis and secretion is also shown. Note, replicated progeny HBV DNA can return to the nucleus by way of an intracellular pathway, which may result in an “auto” amplification of cccDNA.
Attachment and entry
HBV infection of target cells begins with the interactions between viral envelope proteins and its specific cellular receptor(s) on plasma membrane of hepatocytes. While the identity of the cellular receptor(s) for HBV is unknown, the N-terminal region of large envelope protein (called “pre-S1”, see Fig. 1B) appears to be essential in mediating receptor binding and initiating infection [21]. A peptide derived from pre-S1 region potently inhibits HBV infection of HepaRG cells [22, 23], which is evidence for the role of preS1 in HBV binding as well as an example of a possible therapeutic strategy.
Binding to specific cellular receptor(s) is thought to trigger the entry of virion into hepatocyte via endocytosis [24]. Presumably, viral capsid is released into the cytoplasm from endosome upon the fusion of viral envelope and endosomal membranes, a process that might be triggered by low pH and/or proteolytic cleavage of envelope protein [25].
Translocation of the viral rcDNA to the nucleus
Once in the cytoplasm, the capsid delivers its relaxed circular DNA (rcDNA), shown in Fig. 1, that is contained within its virion into the nucleus through nuclear pore complex (NPC) [26]. It has been shown that the intra-cytosolic transportation of capsid is facilitated by its interaction with cellular microtubules [27]. Passage through the NPC is mediated by the interaction between a nuclear localization signal (NLS) on the C termini of the viral capsid proteins and nuclear import receptors importin α and β [28]. Nuclear localization also appears to be cell-cycle dependent [29]. It is assumed that the exposure of NLS is regulated and dependent on genome maturation that induces structural changes of capsid. There is evidence suggesting that the complete disassembly of capsids and release of viral genomic DNA into nucleus most possibly occurs in the nuclear basket of NPC [27].
cccDNA formation and maintenance
cccDNA, as shown in Fig. 3, is the covalently closed circular (ccc) form of the viral chromosome and is essential to the life cycle and maintenance of the infection. Several viral and presumably host functions are required to generate cccDNA from the infecting viral DNA.
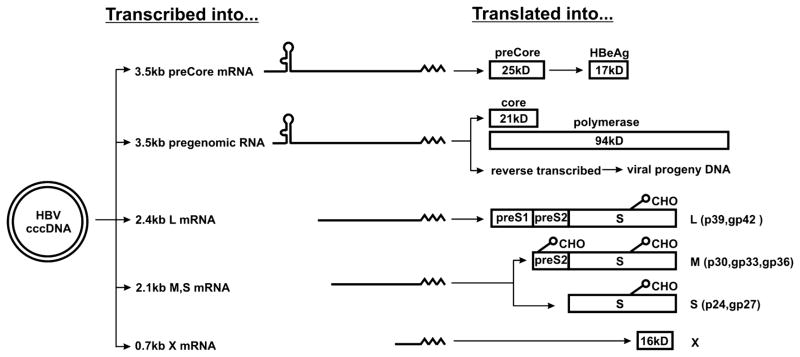
Figure 3. Transcript products of HBV, with the proteins they specify (for mRNA) and encapsidation and replications (for pregenomes).
CHO indicates N-linked glycosylation, and is positioned at the appropriate glycosylation sites. cccDNA, represented by the double lined circle on the left, in transcribed into at least 5 RNA gene products that serve as either pregenomes for encapsidation and/or mRNA molecules that are translated into the proteins indicated. In the case of preCore, it is proteolytically cleaved (by presumably host proteases) into HBeAg. Approximate molecular weight of the (unglycosylated) polypeptide products is indicated in kilodaltons (kD).
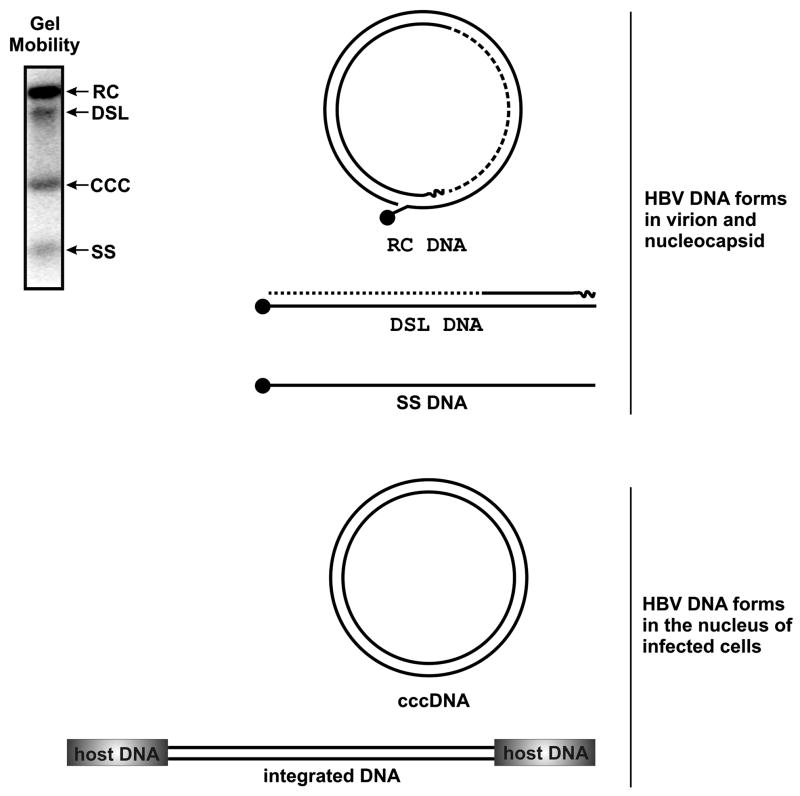
Upon arriving at the nucleus, viral rcDNAs, which are incomplete circles, are converted into a covalently closed circular form (cccDNA). Hepadnavirus rcDNA has several unique structural features. First, as indicated in Fig. 1 & 4, the two strands of viral DNA are asymmetric in length. While the minus strand DNA (the strand that is complementary to the RNA) is the length of one complete genome (unit length), the plus strand DNA is variable in length and usually only approximately 50% of a full length strand [30]. The implication of this is that the strands must be completed early for productive infection to proceed. Secondly, the viral DNA polymerase protein is covalently linked to the 5’ end of minus strand DNA [7]. Thirdly, a capped RNA oligomer of approximately 18 nt in length is linked to the 5’ end of plus strand DNA. This RNA oligomer is derived from 5’ end of pgRNA and serves as a primer for plus strand DNA synthesis [31].
Figure 4. DNA structures associated with HBV.
Relative mobility of the various structures as they appear resolved through agarose gels (upper left), the illustration of their structures and the step in the life cycle with which they are associated.
Conversion of viral rcDNA into cccDNA requires the completion of plus strand DNA synthesis, removal of the terminal modifications and covalent ligation of the ends of both DNA strands. How these biochemical reactions are achieved remains to be resolved. While it is reasonable to believe that general cellular DNA repair enzymes may catalyze the reactions of removal of RNA primer from 5’ end of plus strand DNA and ligation of the two ends of both DNA strands, the removal of DNA polymerase protein from the 5’ end of minus strand DNA represents a very unusual, if not unique, reaction.
Biochemically, this deproteinization reaction can be achieved via either nucleolytic cleavage of viral DNA close to the 5’ end by an endonuclease or hydrolysis of the phosphodiester bond between tyrosine of polymerase and the 5’-phosphoryl group of minus strand DNA. The role of cellular and/or viral proteins, such as DNA polymerase itself, in this reaction remains to be determined. Also, it is not yet known if the deproteinization of rcDNA occurs before or after the translocation of the rcDNA across the nuclear membrane. Knowing this could help in the pursuit of the cellular functions involved in mediating the deproteinization and possibly in fashioning new antiviral strategies.
Unlike other retroviruses, the integration of HBV DNA into host cellular chromosomes is not required for its replication and the cccDNA exists in the nucleus of infected cells as an episomal minichromosome [32]. Nevertheless, the HBV genome can, and frequently does, randomly integrate into cellular chromosomes [33]. The role of HBV DNA integration in oncogenic transformation of infected hepatocytes has been extensively studied and will be discussed below. Interestingly, integrated viral DNA has been used as a genetic marker to monitor virally infected hepatocyte turnover, and it has demonstrated that extensive killing of infected hepatocytes occurs during the resolution of transient WHV infection of woodchucks [34].
cccDNA can apparently persist within the nucleus of an infected hepatocyte for the lifetime of the hepatocyte. It is thus stable, although the degree to which it becomes mutated and repaired is not known. Since cccDNA is the source of progeny viral templates, and viral mutants are a problem in the pathogenesis and therapeutic management of infection, understanding the mutation and repair of cccDNA has clinical implications.
Following cell division, cccDNA may be asymmetrically distributed to progeny cells, and this dilution effect can be a means of its elimination [35]. Otherwise, it appears to remain in the infected cell for months, or even years, in the absence of productive replication of the virus, such as under the treatment of potent viral DNA polymerase inhibitors. The significance of its durability is reflected by the observation that after cessation of antiviral therapy, viral rebound often occurs, and the molecular source of the rebounding viral progeny is surviving cccDNA. Thus, the elimination of cccDNA remains a therapeutic challenge.
Transcription of viral RNA
Once formed, the cccDNA in the nucleus serves as the template for the transcription of four groups of viral RNA, as illustrated in Fig. 3. These are: 3.5kb pre-core mRNA (pre-C) and pregenomic (pg) RNAs, 2.4kb mRNA for large (L) envelope protein; 2.1kb mRNA for middle (M) and major surface (S) proteins and 0.7kb mRNA for the X protein.
Pre-C mRNA is translated to produce a “pre-C” protein that is further proteolytically processed into e antigen (HBeAg). HBeAg is not a component of viral and subviral particles, but is secreted from infected cells and its detection in the circulation (by commercial assays) is a surrogate marker for high levels of viral replication. As stated above, HBeAg is a product of the pre-C gene and transcript (Fig. 3), that is derived directly from cccDNA. Hence, the level of HBeAg in the circulation correlates, in general, with viremia (viral DNA levels in the circulation) (5). Loss of detectable HBeAg in individuals with chronic viral hepatitis and the appearance of detectable levels of antibody to HBeAg is usually considered to be a beneficial milestone and evidence of reduced viral replication [36]. However, a significant fraction of chronic HBV carriers who appear to lose HBeAg have done so because of mutations in pre-C region, and these HBeAg mutant viruses may even be more pathogenic than are the wild type [37]. Thus, loss of HBeAg alone, particularly when viral DNA levels remain high, can not be taken as a favorable sign. Curiously, the biological function of HBeAg remains a mystery, although it may be important for natural infection [1, 38] and a role in immunosuppression has been proposed [11].
A further complication is the question as to whether or not pharmacological induction of HBeAg negativity and natural reduction, are biologically equivalent. Since natural reduction almost certainly means there has been a degree of immunological arousal, and pharmacological reduction of viral levels does not require this, it seems likely that the two pathways leading to eAg reduction are quite different. Having said that, since HBeAg may be actively opposing immune recognition to HBV [11], reducing HBeAg, even indirectly, by inhibiting DNA synthesis, may allow for immune mechanisms to get “jump started” and play in role in the durable antiviral suppression often seen in those with antiviral induced HBeAg conversions.
Besides serving as mRNA for viral capsid protein and DNA polymerase, pgRNA serves as the template for the reverse transcriptional synthesis of viral DNA [44]. Viral DNA cis-elements and cellular transcription factors that direct and regulate viral RNA transcription has been extensively studied, and comprehensive reviews on this subject have been published [45, 46].
The transcripts of HBV and related animal hepadnaviruses do not usually contain introns, which are segments of RNA, present in primary, pre-mRNA transcripts, that are excised and not present in the mRNA, created by splicing the remaining RNA together, as do most mammalian pre-mRNA. However, spliced viral RNAs have been reportedly detected in transfected cells and virally infected livers. In most cases, the roles of those spliced RNA in viral replication have not been established. A spliced form of the duck HBV pgRNA, has been reported to serve as a second mRNA for large envelope protein, and may be essential for viral particle secretion from infected primary duck hepatocytes [47].
Synthesis of progeny viral DNA genomes from pgRNA in the cytoplasmic capsids
A unique property of HBV replication is that viral DNA replication occurs inside immature nucleocapsids via protein-primed reverse transcription as illustrated in Fig. 2. Briefly, the DNA polymerase protein binds to the pgRNA at its 5’ epsilon stem-loop structure to initiate nucleocapsid assembly and prime viral minus-strand DNA synthesis to extend for three nucleotides. Subsequently, polymerase and covalently attached nascent DNA is translocated to the 3’ copy of direct repeat 1 (DR1), and minus strand DNA synthesis continues by copying pgRNA [7, 8, 48]. The latter is degraded by an RNase H activity of polymerase during minus strand DNA synthesis. When the polymerase reaches the 5’ end of pgRNA, an RNA oligomer containing DR1 and the 6 to 7 nucleotides at the 5’ end of DR1 sequence is left uncleaved. It is this RNA oligomer that is subsequently translocated and annealed to the direct repeat 2 (DR2) and primes plus strand DNA synthesis to yield relaxed circular DNA (rcDNA) [31].When rc DNA are formed, the nucleocapsids are matured and can be enveloped and secreted out of cells [49, 50].
An alternative to being enveloped and exported as virion is for newly formed mature capsids containing rcDNA to be “shunted” into the nucleus where they are presumably converted into more cccDNA molecules [51]. Thus, this is a way in which cccDNA levels can be increased in the absence of re-infection of cells. This intracellular amplification pathway operates very efficiently in the early stage of viral infection to build up a nuclear cccDNA pool. A series of elegant studies reported by Summers and colleagues suggested that DHBV large envelope protein (L) regulated intracellular DNA traffic and cccDNA formation [52, 53]. At the early time of infection, when L protein is at low level, capsid nuclear delivery is favored and allows accumulation of cccDNA which ensures that the infected cells will be stably colonized. At the later stage of infection, large amounts of protein are made and capsid will be enveloped and secreted out of cells.
Assembly of viral particles and secretion
Virions are assembled on, and bud from, endoplasmic reticulum (ER) membrane through the engulfing of rc or dsl DNA containing capsids (so-called mature capsids) in ER membrane studded with viral envelope proteins. As mentioned previously, HBV encodes three envelope proteins called Large (LHBs), Medium (MHBs) and Small (SHBs) surface antigens. They are all synthesized from the same open reading frame of the viral genome by using different starting codons (Fig. 3). Thus, they all share a common C-termini and SHBs domain. The LHBs and MHBs proteins contain SHBs plus amino-terminal extensions of pre-S2, pre-S1 and pre-S2 domains, respectively.
As illustrated in Fig. 3, SHBs is monoglycosylated and is the major viral surface protein. Virions contain approximately 100 copies of SHBs for every 5 MHBs, and 1 LHBs, protein. Moreover, as mentioned in the beginning of this review, an infected cell typically secretes hundreds to thousands of polymers of 22 nm particles of SHBs devoid of capsid and nucleic acids for every virion. Indeed the serum of an individual chronically infected with HBV typically contains hundreds of micrograms per ml of empty SHBs particles, which are essentially lipoprotein polymers of sHBS containing a smaller amount of MHBs. It is thus SHBs that is usually measured as “HBsAg” by commercial assays, and its consistent detection in the circulation in people over a time of more than six months is considered as evidence of chronic infection. Although it is possible that it could be produced from defective viral genomes integrated in host cellular chromosomes, and would thus be stably produced in the absence of productive viral infection, this is considered to be very unusual.
Nascent viral envelope polypeptides are very quickly oligomerized. We have speculated that their rapid oligmerization in the ER makes them very refractory to degradation by cellular proteasomes [54]. Since degradation by cellular proteasomes provides peptides for presentation to cytotoxic T lymphocytes by MHC class 1 molecules, resistance to proteasome degradation may provide a means by which HBV can remain “invisible” to the host immune system thereby contributing to the establishment of chronic infection. This notion will be discussed in more detail below.
As mentioned above, pre-S1 domain of LHBs is believed to mediate the binding of virions to its cellular receptor(s). This function requires the pre-S1 domain of the protein to be exposed on the outside of viral envelope membrane. But surprisingly, it has been shown that for a fraction (approx. 50%) of LHBs protein in virions, their pre-S1 domains are projected into the interior of viral envelope [55–57]. Further studies on the topology of LHBs on ER membrane revealed that the observed dual topology of LHBs is originated from the partial membrane translocation of their pre-S domains, which probably occurs during viral particle assembly and budding from ER membrane into ER lumen [58]. It has been shown that the cytosolic deposition of pre-S1 domain provide docking sites for matured viral capsids and is essential for virion assembly and budding. In this sense, the pre-S domain functions as a matrix protein [56].
Additionally, LHBs is modified by N-terminal myristylation [59]. This post-translational modification has been shown to be dispensable for virion assembly, but essential for infectivity. M protein is absent in DHBV. MHBs is not required for HBV virion secretion, and their biological function is not well understood.
In contrast to most retroviruses, hepadnaviruses contain rcDNA genomes in their infectious viral particles (virions). This is apparently because the viral pgRNA containing capsids cannot be enveloped and secreted from infected cells [60]. A hypothesis that is supported by the results obtained from both genetic and kinetic studies is that capsid maturation signals exist that confer the ability of capsids to interact with the pre-S1 domain of large envelope proteins on ER membrane and initiate virion assembly and budding [49]. Thus far, the details of the maturation signals remain to be identified.
It has been shown that while intracellular capsid protein in DHBV infected cells is heterogeneously phosphorylated, the capsid protein in secreted DHBV virions is dephosphorylated [61]. It has therefore been suggested that capsid protein dephosphorylation may be triggered by the formation of at least partially double-stranded DNA inside capsids and serve as a maturation signal. However, given the large excess of empty capsids in viral infected cells, this hypothesis is difficult to be proven experimentally. Alternatively, a structural change of capsid, which can be triggered by the synthesis of mature rcDNA, could also expose the pre-S1 interacting domain of capsid protein and thus serve as a maturation signal.
Immunological basis of chronicity
Despite the fact that most adulthood HBV infections are transient, approximately 1 to 5 % of people infected as adults and more than 90% of those infected as neonates fail to mount a sufficient immune response to clear the virus, and develop a life-long chronic infection. Resolution of acute infection is associated with a vigorous polyclonal helper (Th) and cytotoxic T lymphocyte (CTL) response to multiple viral antigens in the infected livers [2, 10, 62, 63]. Moreover, although destruction of virally infected hepatocytes is evident during the resolution of acute infection, it has been elegantly demonstrated that the noncytolytical reduction of viral gene products in infected cells by cytokines, such as interferon gamma and tumor necrosis factor, released from activated T lymphocytes is likely to play an important role in terminating the infection [12, 64].
In marked contrast to those achieving resolution of acute infection, chronic carriers of HBV appear to have an inadequate (in quality and quantity) T cell response to viral antigens [12, 63–66]. The reason for the failure of mounting a sufficient immune response against the virus in chronic carriers still remains a mystery. It is reasonable to anticipate that the outcome might be determined during the early phase of infection. In this period of time, there is a growing antigenemia with the cellular immune response, curiously, lagging behind. Furthermore, although infection with hepatitis C virus (HCV) has been shown to induce a myriad of host innate response genes [67], HBV infection does not activate a detectable host cellular innate immune response as determined by microarray analysis of transcriptomes of HBV infected chimpanzee livers [68]. The reason that HBV can “fly under the radar screen” is not known. Hence, there may be distinct mechanisms whereby HBV actively inhibits or avoids innate and adaptive immune responses, which contribute to tip the balance between resolution and chronicity [68].
One simple explanation for the observed inadequate immune CTL response in chronic HBV carriers is of great interest and suggests a potential approach for the activation of antiviral cellular immunity by therapeutic vaccination. As mentioned above, MHC I presentation of viral epitopes depends upon the degradation of viral proteins by cellular proteasomes. Thus, processes that inhibit the degradation of viral proteins would be expected to inhibit the ability of the circulating immune system to recognize infected cells. Viral protein complexes that form within the ER, such as in the case of HBV envelope proteins would be too large to be retrotranslocated from the ER to the cytosolic proteasomes. HBsAg particles that have formed immediately following synthesis would certainly be too large to pass through translocon pores that may have limits of less than 100 angstroms. Moreover, since they are lipoproteins within vesicles, resistance to ER associated degradation is all the greater. Thus, this may be an explanation for the relatively poor CTL response to the HBV envelope proteins, either in a natural setting [63, 65] or with DNA vaccines [69]. In this scenario, early in infection, the virus would have a chance to establish itself by resisting detection by the circulating immune system.
Consistent with this notion, Mehta and colleagues reported that expression of mutant HBsAg molecules that fail to be translocated into ER and thereby degrade very quickly in cytoplasm are much better inducers of CTL response in cultured cells [70]. It remains to be seen if DNA-based vaccination with the same expression constructs will induce a stronger CTL response in animals and break the apparent immune tolerance in chronic HBV carriers.
Molecular biology of the oncogenesis of HCC in chronic HBV infection
Hepatocellular carcinoma (HCC) is the 5th most common cancer, but the 3rd leading cause of cancer death in the world with more than 500,000 fatalities annually. The major etiology of HCC/liver cancer in people is HBV followed by HCV infection, although non-viral causes also play a role in a minority of cases.
The molecular etiology of HBV induced HCC remains, for the most part, unclear. It is worth pointing out that there have been reports of HBV DNA integrations near cellular proto-oncogenes in tumor cells. Although the insertion activation of oncogene expression may indeed occur, it is unusual in people and may be responsible for only a minority of HBV induced HCC [71]. Thus, there has been great interest in determining if any of the viral gene products and host factors is responsible for malignant transformation.
Role of X protein
The 154 amino acid viral gene product “X protein” is probably the viral function most frequently implicated in oncogenesis. It is named “X protein” or HBx because of the uncertainty about its function during a natural viral infection. It has been shown that WHV X protein is not essential for WHV DNA replication in cultured cells, but is absolutely required for its infectivity in vivo [72]. X transgenic mice have been reported to be predisposed to development of liver cancer, heightening interest in a role for “X” in oncogenesis [73, 74]. Although there are many theories, there is no current consensus on if or how “X” mediates HBV associated cancers.
The “X” protein has also been reported to “trans-activate” numerous cellular gene functions including c-fos, c-jun, c-myc, EGF and MHC I as well as viral genes [75]. Because HBx is not a DNA binding protein, its transactivation function must be due to its interaction with other cellular proteins. Thus far, it has been reported that HBx can interact with several well-known signaling proteins such as PI3K p85 subunit, which is a component of PI3K-Akt signaling pathway as well as p53 and even proteins involving in DNA damage repair and degradation [76–78].
Taken together, given “X”s apparent ability to influence many functions associated with cellular transformation, speculation of its central role in oncogensis remains high. As of the drafting of this manuscript, however, its specific role, if any, remains the subject of debate. The transcripts associated with each gene product are provided in the figures.
Roles of other viral proteins
HBV envelope glycoproteins are also associated with transformation and HBV induced diseases. Specifically, accumulation of large envelope protein (LHBs) within the ER of hepatocytes has been shown to be associated with predisposition to transformation in transgenic animal studies and with severe fulminant hepatitis in people [79]. Furthermore, previous studies showed that integrated HBV DNA in the human hepatoma cell line Huh4 encodes a carboxyl terminally truncated MHBs protein, so called MHBst [80, 81]. It has been demonstrated that this variant M protein activates c-raf-1/Erk2 signal transduction pathway in transgenic mice and leads to increased hepatocyte proliferation and higher incidence of liver tumors [82]. But thus far, the role of truncated envelope protein variants in hepatocarcinogenesis of HBV infection in human beings remains unknown.
Role of host factors
Although it seems likely that viral gene products can predispose the transformation, as explained above, no HBV specified oncogene has been clearly implicated. Thus, it is possible that hepatocarcinogenesis in chronic HBV infection arises most often from a combination of viral induced host factors and the establishment of an environment that is permissive for oncogenic transformation. For example, oncogenic transformation may result from liver damage by way of CTLs attacking infected hepatocytes and inducing constant liver cell regeneration. The constant regeneration increases the chances of errors and mutations in the dividing liver cells, and the chances of error are increased further when the reservation occurs in an environment rich with free radicals and other mutagens resulting from an inflammatory environment.
It is worth noting that, compared to uninfected individuals, the half life of hepatocytes is much shorter [1, 83]. The environment in which the hepatocytes replicate in response to injury is likely to be enriched in mutagens such as oxidants, which are present during, and as a consequence of, cell injury and immunological attack. These complications are all aggravated during fibrosis, which is also a predisposition for liver cancer.
Immunization
Antibodies against the “a” epitope of the HBs envelope protein are protective [84]. Their appearance following natural infection is associated with resolution, and their elicitation following immunization is the basis of protection (80). Initially, subviral particles devoid of viral nucleic acid, purified from the circulation of chronic HBV carriers, were used as a vaccine (1, 10, 11, 80). It was shown to be efficacious, and despite great efforts to ensure that all infectious agents that might co-purify with the subviral particles were inactivated, concerns regarding the possibility of contamination have caused this first generation of vaccine to go into disuse in the United States.
Current HBV vaccines, in use in North America, are purified subviral particles produced from yeast expressing recombinant genes. These vaccines are efficacious and safe, and a series of three injections can apparently confer long-term protective immunity [85–87]. Whether or not protection conferred by injection of the subunit vaccine is lifelong is not yet known. This could be a special concern for those vaccinated at very early ages. It would be unusual, for example, for a polypeptide vaccine, given in infancy, to confer life long immunity. The role of T cells in contributing to protection under these circumstances is not clear, but levels of humoral antibody above a minimum are considered essential for protection.
Phenotype and genotypes defined by sHBS and the genome
sHBS carries the “a” antigenic determinant, created by two “loops” of amino acids 120–163, that is recognized by most commercial assays and the epitope to which neutralizing antibody is raised [84, 88, 89]. This is the major group, serological antigenic determinant, although at least two other sub-dominant determinants have also been identified with the second being either “d” or “y” and the third being either “w” or “r.” Hence, except for “a” mutants (see below), the major subtypes of HBV are phenotypically defined serologically and comprised of the “a” determinant and two other sub-dominant determinants, and hence called “adw”, “adr”, “ayw” or “ayr.” Genotypes of the HBV virus are designated by capital letters A, B, C, D – H. Genotypes are defined as having a greater than 8% non homology from Genotype A.
The recombinant vaccines currently used elicit antibodies against only, or mostly, a single epitope within the HBsAg called “a.” Thus, the possibility exists that vaccine escape mutants, which have mutated amino acids within this epitope and are not recognized by anti- “a” epitope antibody, can emerge. This could become a problem over time, especially where the vaccine is administered to an infant born from mothers with high titers of virus, since the possibility of pre-existing mutants would be high.
The American Committee on Immunization Practices’ (ACIP) recommendations call for administration of HBV vaccine and passive immunoglobulin to infants born from HBV infected mothers in the delivery room [85–87]. Remarkably, this post exposure intervention is highly effective in preventing establishment of chronic infection in the newborn. However, in circumstances where hepatitis B immune globulin (HBiG, HBIG) is not co-administered, due to noncompliance, limited resources in the developing world or perhaps viral titer is so high as to be unmanageable, it is not difficult to see how vaccine escape mutants could emerge. A similar situation can be imagined for HBV chronic carriers receiving liver transplantation supported by HBiG. Indeed, there are now several reports of the surprising prevalence of HBV “a” antigen mutants, which would be expected to refractory to current “a” epitope dependent vaccines [90]. In one report, the incidence of HBsAg mutants in infants born from HBV carrier mothers and treated with vaccine and HBiG was 4% [43]. Thus, development of novel vaccines that induce antibodies against other potential epitopes, such as epitopes located in pre-S1 and pre-S2 region, is warranted.
Therapeutic intervention, the molecular perspective
Currently available antiviral treatment of chronic hepatitis B includes alpha interferon (IFN-α) and four nucleotide or nucleoside analogues. It has been shown that IFN-α could potently inhibit HBV replication by preventing pgRNA containing nucleocapsid assembly in a mouse hepatocyte-derived cell line, and in HBV transgenic mice [68, 91]. But ironically, IFN-α only modestly inhibits HBV replication in human hepatoma cell lines [92]. Thus, HBV does certainly appear to have some level of sensitivity to IFN-α, although this sensitivity may be genotype specific, with genotype C more resistant than the others. It has been reported that a forty-eight week treatment of peginterferon alpha 2a can result in HBeAg serum conversion and reduction of viral load in 30–40% of the patients tested [42, 93]. Nevertheless, the therapeutic value of IFN-α is important, and there have even been reports of statistically significant numbers of chronic carriers who are treated with the cytokine and reach the important milestones of HBsAg loss as well with detectable levels of anti-HBs [41, 93].
There are currently three nucleoside analogues (Epivir/Lamivudine, Entecavir/Baraclude, Telbivudine/Tyzeka) and one nucleotide analogue (Adefovir/Hepsera) that have been approved by FDA for the therapy of chronic hepatitis B. Those drugs selectively inhibit HBV DNA replication by causing premature chain termination. Despite the potent inhibition of viral replication, even prolonged nucleoside analogue treatments rarely cure HBV infection, and virological relapse is common following the discontinuation of therapy [40, 41, 94]. In addition, the development of drug resistance to the polymerase inhibitors by the virus will most likely limit their long-term efficacy [41]. As mentioned previously, the major reason for limitations in the durable elimination of chronic HBV infection with the HBV DNA polymerase inhibitors may be that the drugs do not necessarily eliminate the cccDNA.
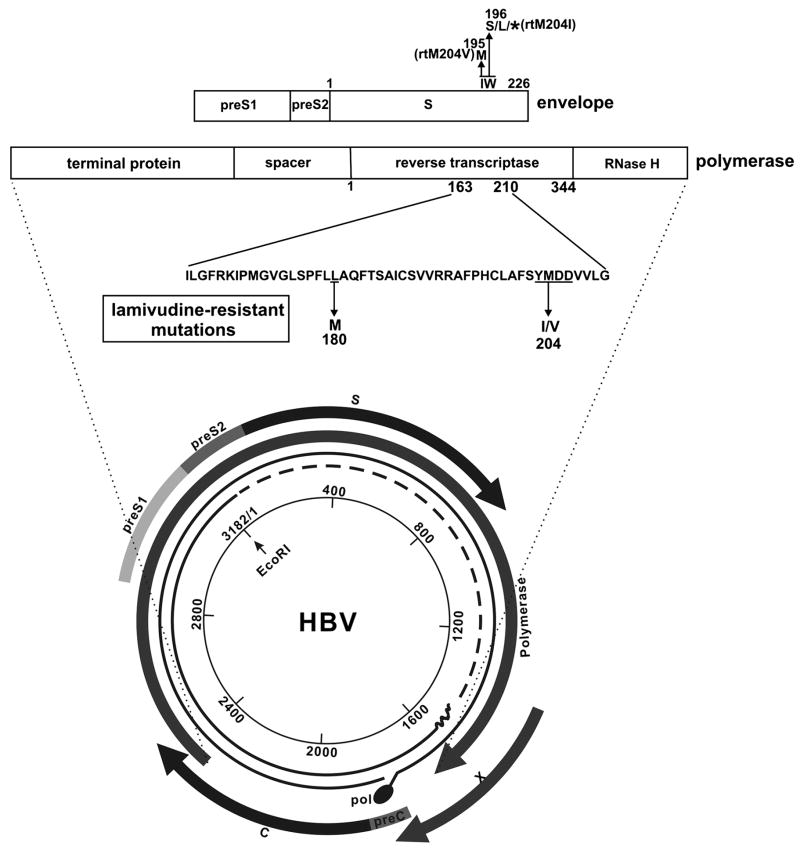
One complicating but potentially very important point is that, although all transcripts come from the same DNA strand, all reading frames are used, and the genes for several of the different HBV functions overlap. For example, as shown in Fig. 5, the gene for the polymerase overlaps with the envelope and X genes. The gene for X overlaps with the beginning of the pre-Core transcript. Thus, mutations in one gene may affect the coding specificity of another. Of particular note is that mutations within the polymerase gene (some of which have been associated with resistance to antiviral agents such as lamivudine), also alter the amino acids within the envelope proteins . This is illustrated in the Fig. 5. The biological significance of this is uncertain, although in some cases changes within epitopes recognized by the cellular immune system have been observed [95], and is reasonable to assume that immunological recognition of therapy induced epitopes could provide an additional, unexpected, influence upon outcome.
Figure 5. Mutations within the pol gene can result in changes within the envelope protein.
The genome HBV is shown, as in Fig. 1, as a circle, with 1 to 3,182 nucleotides numbered consecutively from the single Eco R1 site. The coding regions for each gene are shown as curved arrows that follow the direction of the transcripts. The polypeptide specified by the pol (polymerase) and env (envelope) genes are expanded as horizontal boxes. The amino acid sequence of the polymerase between amino acids 163 and 210 is the region that contains amino acids that, when mutated, can confer resistance to some polymerase inhibitors. The YMDD amino acid motif (underlined), for example, when mutated (as shown) confers resistance to lamivudine and is provided with the amino acid transition indicated. Since the open reading frames for pol and env overlap, (amino acid 204 of the rt (reverse transcriptase) roughly correspond to amino acid 195/6 of the envelope protein, as indicated) a change in this motif will change the amino acids specified by the env open reading frame, and this is concomitant change is indicated.
Additionally, several reports have indicated that assembly of capsids and packaging of pgRNA into capsids can be efficiently inhibited by small molecules in cultured cells and HBV transgenic mice in vivo [41, 96–98]. Further exploration of those classes of compounds and the new viral targets should lead to the development of novel antiviral drugs that can be used in combination with currently available DNA polymerase inhibitors to enhance their antiviral efficacy and reduce the chance of the appearance of drug-resistant HBV mutants, which is a major limitation of long-term DNA polymerase inhibitor monotherapy.
Conclusion
Elimination of cccDNA from infected hepatocytes and re-establishment of host antiviral immune response against HBV are important therapeutic objectives. Future studies of HBV biology should reveal the molecular mechanisms of infectious virus entry into hepatocytes and molecular pathway of cccDNA formation, and may provide clues regarding ways in which cccDNA can be reduced. HBV chronicity and oncogenesis, itself, are almost certainly the result of a cat and mouse game in which virus mediated immuno-evasion and persistent replication is countered by incomplete host immuno-destruction. No single viral or host factor is responsible for this balancing act, but it is clear that chronically infected individuals are not immuno-indolent, and that the virus exerts far greater control over its immuno-invisibility than perhaps previously thought. The more that is learned about these details, the more likely is the development of new biomarkers of disease detection and therapeutic agents.
Acknowledgments
The authors received support from the Hepatitis B Foundation, the Commonwealth of Pennsylvania and The National Institute of Allergy and Infectious Diseases of the US National Institutes of Health in preparation of this manuscript. Ms. Erica Litschi is thanked for her help in manuscript preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Seeger C, Mason WS. Hepatitis B virus biology. Microbiology & Molecular Biology Reviews. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganem D, Prince AM. Hepatitis B infection: natural history and clinical consequences. New England Journal of Medicine. 2004;350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 3.Menne S, Tennant BC. Unraveling hepatitis B virus infection of mice and men (and woodchucks and ducks) Nature Medicine. 1999;5:1125–1126. doi: 10.1038/13445. [DOI] [PubMed] [Google Scholar]
- 4.Guo H, Mason WS, Aldrich CE, Saputelli JR, Miller DS, Jilbert AR, Newbold JE. Identification and characterization of avihepadnaviruses isolated from exotic anseriformes maintained in captivity. J Virol. 2005;79:2729–2742. doi: 10.1128/JVI.79.5.2729-2742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoofnagle JH, di Bisceglie AM. The treatment of chronic viral hepatitis. New England Journal of Medicine. 1997;336:347–356. doi: 10.1056/NEJM199701303360507. [DOI] [PubMed] [Google Scholar]
- 6.McMahon BJ. Epidemiology and natural history of hepatitis B. Seminars in Liver Disease. 2005;25:3–8. doi: 10.1055/s-2005-915644. [DOI] [PubMed] [Google Scholar]
- 7.Wang GH, Seeger C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell. 1992;71:663–670. doi: 10.1016/0092-8674(92)90599-8. [DOI] [PubMed] [Google Scholar]
- 8.Wang GH, Seeger C. Novel mechanism for reverse transcription in hepatitis B viruses. J Virol. 1993;67:6507–6512. doi: 10.1128/jvi.67.11.6507-6512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu J, Toft DO, Seeger C. Hepadnavirus assembly and reverse transcription require a multi- component chaperone complex which is incorporated into nucleocapsids. Embo J. 1997;16:59–68. doi: 10.1093/emboj/16.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milich DR. Pathobiology of acute and chronic hepatitis B virus infection: an introduction. Journal of Viral Hepatitis. 1997;4:25–30. doi: 10.1111/j.1365-2893.1997.tb00177.x. [DOI] [PubMed] [Google Scholar]
- 11.Milich DR, Liang TJ. Exploring the biological basis of hepatitis B e antigen in hepatitis B virus infection. Hepatology. 2003;38:1075–1086. doi: 10.1053/jhep.2003.50453. [DOI] [PubMed] [Google Scholar]
- 12.Guidotti LG, Rochford R, Chung J, Shapiro M, Purcell R, Chisari FV. Viral clearance without destruction of infected cells during acute HBV infection. Science. 1999;284:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]
- 13.Walter E, Keist R, Niederost B, Pult I, Blum HE. Hepatitis B virus infection of tupaia hepatocytes in vitro and in vivo. Hepatology. 1996;24:1–5. doi: 10.1002/hep.510240101. [DOI] [PubMed] [Google Scholar]
- 14.Jilbert AR, Miller DS, Scougall CA, Turnbull H, Burrell CJ. Kinetics of duck hepatitis B virus infection following low dose virus inoculation: one virus DNA genome is infectious in neonatal ducks. Virology. 1996;226:338–345. doi: 10.1006/viro.1996.0661. [DOI] [PubMed] [Google Scholar]
- 15.Jilbert AR, Botten JA, Miller DS, Bertram EM, Hall PM, Kotlarski J, Burrell CJ. Characterization of age- and dose-related outcomes of duck hepatitis B virus infection. Virology. 1998;244:273–282. doi: 10.1006/viro.1998.9095. [DOI] [PubMed] [Google Scholar]
- 16.Paran N, Geiger B, Shaul Y. HBV infection of cell culture: evidence for multivalent and cooperative attachment. Embo J. 2001;20:4443–4453. doi: 10.1093/emboj/20.16.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acs G, Sells MA, Purcell RH, Price P, Engle R, Shapiro M, Popper H. Hepatitis B virus produced by transfected Hep G2 cells causes hepatitis in chimpanzees. ProcNatlAcadSci USA. 1987;84:4641–4644. doi: 10.1073/pnas.84.13.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sureau C, Eichberg JW, Hubbard GB, Romet-Lemonne JL, Essex M. A molecularly cloned hepatitis B virus produced in vitro is infectious in a chimpanzee. JVirol. 1988;62:3064–3067. doi: 10.1128/jvi.62.8.3064-3067.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sureau C, Romet-Lemonne J, Mullins JI, Essex M. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with cloned circular HBV DNA. Cell. 1987;47:37–47. doi: 10.1016/0092-8674(86)90364-8. [DOI] [PubMed] [Google Scholar]
- 20.Gripon P, Rumin S, Urban S, Le Seyec J, Glaise D, Cannie I, Guyomard C, Lucas J, Trepo C, Guguen-Guillouzo C. Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci U S A. 2002;99:15655–15660. doi: 10.1073/pnas.232137699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tai PC, Suk FM, Gerlich WH, Neurath AR, Shih C. Hypermodification and immune escape of an internally deleted middle-envelope (M) protein of frequent and predominant hepatitis B virus variants. Virology. 2002;292:44–58. doi: 10.1006/viro.2001.1239. [DOI] [PubMed] [Google Scholar]
- 22.Urban S, Gripon P. Inhibition of duck hepatitis B virus infection by a myristoylated pre-S peptide of the large viral surface protein. Journal of Virology. 2002;76:1986–1990. doi: 10.1128/JVI.76.4.1986-1990.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gripon P, Cannie I, Urban S. Efficient inhibition of hepatitis B virus infection by acylated peptides derived from the large viral surface protein. Journal of Virology. 2005;79:1613–1622. doi: 10.1128/JVI.79.3.1613-1622.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmitt S, Glebe D, Alving K, Tolle TK, Linder M, Geyer H, Linder D, Peter-Katalinic J, Gerlich WH, Geyer R. Analysis of the pre-S2 N- and O-linked glycans of the M surface protein from human hepatitis B virus Structure and glycosylation patterns of surface proteins from woodchuck hepatitis viru. J Biol Chem. 1999;274:11945–11957. doi: 10.1074/jbc.274.17.11945. [DOI] [PubMed] [Google Scholar]
- 25.Lu X, Block TM, Gerlich WH. Protease-induced infectivity of hepatitis B virus for a human hepatoblastoma cell line. J Virol. 1996;70:2277–2285. doi: 10.1128/jvi.70.4.2277-2285.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kann M, Schmitz A, Rabe B. Intracellular transport of hepatitis B virus. World J Gastroenterol. 2007;13:39–47. doi: 10.3748/wjg.v13.i1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreyav HJN, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N, Beranek M, Jandik P, Benamouzig R, Jullian E, Laurent-Puig P, Olschwang S, Muller O, Hoffmann I, Rabes HM, Zietz C, Troungos C, Valavanis C, Yuen ST, Ho JWC, Croke CT, O'Donoghue DP, Giaretti W, Rapallo A, Russo A, Bazan V, Tanaka M, Omura K, Azuma T, Ohkusa T, Fujimori T, Ono Y, Pauly M, Faber C, Glaesener R, de Goeij A, Arends JW, Andersen SN, Lovig T, Breivik J, Gaudernack G, Clausen OPF, DeAngelis P, Meling GI, Rognum TO, Smith R, Goh HS, Font A, Rosell R, Sun XF, Zhang H, Benhatter J, Losi L, Lee JQ, Wang ST, Clarke PA, Bell S, Quirke P, Bubb VJ, Piris J, Cruickshank NR, Morton D, Fox JC, Al-Mulla F, Lees N, Hall CN, Snary D, Wilkinson K, Dillon D, Costa J, Pricolo VE, Finkelstein SD, Thebo JS, Senagore AJ, Halter SA, Wadler S, Malik S, Krtolica K, Urosevic N. Kirsten ras mutations in patients with colorectal cancer: the 'RASCAL II' study. British Journal of Cancer. 2001;85:692–696. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kann M, Sodeik B, Vlachou A, Gerlich WH, Helenius A. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J Cell Biol. 1999;145:45–55. doi: 10.1083/jcb.145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeh CT, Chiu HT, Chu CM, Liaw YF. G1 phase dependent nuclear localization of relaxed-circular hepatitis B virus DNA and aphidicolin-induced accumulation of covalently closed circular DNA. J Med Virol. 1998;55:42–50. doi: 10.1002/(sici)1096-9071(199805)55:1<42::aid-jmv8>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 30.Summers J, O'Connell A, Millman I. Genome of hepatitis B virus: restriction enzyme cleavage and structure of DNA extracted from Dane particles. Proc Natl Acad Sci USA. 1975;72:4597–4601. doi: 10.1073/pnas.72.11.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lien JM, Aldrich CE, Mason WS. Evidence that a capped oligoribonucleotide is the primer for duck hepatitis B virus plus-strand DNA synthesis. Journal of Virology. 1986;57:229–236. doi: 10.1128/jvi.57.1.229-236.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newbold JE, Xin H, Tencza M, Sherman G, Dean J, Bowden S, Locarnini S. The covalently closed duplex form of the hepadnavirus genome exists in situ as a heterogeneous population of viral minichromosomes. Journal of Virology. 1995;69:3350–3357. doi: 10.1128/jvi.69.6.3350-3357.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Block TM, Mehta AS, Fimmel CJ, Jordan R. Molecular viral oncology of hepatocellular carcinoma. Oncogene. 2003;22:5093–5107. doi: 10.1038/sj.onc.1206557. [DOI] [PubMed] [Google Scholar]
- 34.Summers J, Jilbert AR, Yang W, Aldrich CE, Saputelli J, Litwin S, Toll E, Mason WS. Hepatocyte turnover during resolution of a transient hepadnaviral infection. Proc Natl Acad Sci U S A. 2003;100:11652–11659. doi: 10.1073/pnas.1635109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang YY, Zhang BH, Theele D, Litwin S, Toll E, Summers J. Single-cell analysis of covalently closed circular DNA copy numbers in a hepadnavirus-infected liver. Proc Natl Acad Sci U S A. 2003;100:12372–12377. doi: 10.1073/pnas.2033898100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tong QS, Zheng LD, Wang L, Zeng FQ, Chen FM, Dong JH, Lu GC. Downregulation of XIAP expression induces apoptosis and enhances chemotherapeutic sensitivity in human gastric cancer cells. Cancer Gene Therapy. 2005;12:509–514. doi: 10.1038/sj.cgt.7700813. [DOI] [PubMed] [Google Scholar]
- 37.Tong SP, Li JS, Vitvitski L, Trepo C. Active hepatitis B virus replication in the presence of anti-HBe is associated with viral variants containing an inactive pre-C region. Virology. 1990;176:596–603. doi: 10.1016/0042-6822(90)90030-u. [DOI] [PubMed] [Google Scholar]
- 38.Ou JH. Molecular biology of hepatitis B virus e antigen. J Gastroenterol Hepatol. 1997;12:S178–187. doi: 10.1111/j.1440-1746.1997.tb00499.x. [DOI] [PubMed] [Google Scholar]
- 39.Liaw YF. Impact of YMDD mutations during lamivudine therapy in patients with chronic hepatitis B. Antivir Chem Chemother. 2001;12(Suppl 1):67–71. [PubMed] [Google Scholar]
- 40.Lai CL, Rosmawati M, Lao J, Van Vlierberghe H, Anderson FH, Thomas N, Dehertogh D. Entecavir is superior to lamivudine in reducing hepatitis B virus DNA in patients with chronic hepatitis B infection. Gastroenterology. 2002;123:1831–1838. doi: 10.1053/gast.2002.37058. [DOI] [PubMed] [Google Scholar]
- 41.Werle-Lapostolle B, Bowden S, Locarnini S, Wursthorn K, Petersen J, Lau G, Trepo C, Marcellin P, Goodman Z, Delaney WE, Xiong S, Brosgart CL, Chen SS, Gibbs CS, Zoulim F. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology. 2004;126:1750–1758. doi: 10.1053/j.gastro.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 42.Janssen HL, van Zonneveld M, Senturk H, Zeuzem S, Akarca US, Cakaloglu Y, Simon C, So TM, Gerken G, de Man RA, Niesters HG, Zondervan P, Hansen B, Schalm SW. Pegylated interferon alfa-2b alone or in combination with lamivudine for HBeAg-positive chronic hepatitis B: a randomised trial. Lancet. 2005;365:123–129. doi: 10.1016/S0140-6736(05)17701-0. [DOI] [PubMed] [Google Scholar]
- 43.Tabor E. Infections by hepatitis B surface antigen gene mutants in Europe and North America. Journal of Medical Virology. 2006;78:S43–47. doi: 10.1002/jmv.20606. [DOI] [PubMed] [Google Scholar]
- 44.Seeger C, Mason WS. Replication of the hepatitis virus genome. In: DePamphilis ML, editor. In DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1996. pp. 815–831. [Google Scholar]
- 45.Moolla N, Kew M, Arbuthnot P. Regulatory elements of hepatitis B virus transcription. J Viral Hepat. 2002;9:323–331. doi: 10.1046/j.1365-2893.2002.00381.x. [DOI] [PubMed] [Google Scholar]
- 46.Henkler FF, Koshy R. Hepatitis B virus transcriptional activators: mechanisms and possible role in oncogenesis. J Viral Hepat. 1996;3:109–121. doi: 10.1111/j.1365-2893.1996.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 47.Obert S, Zachmann-Brand B, Deindl E, Tucker W, Bartenschlager R, Schaller H. A spliced hepadnavirus RNA is essential for virus replication. EMBO J. 1996;15:2565–2574. [PMC free article] [PubMed] [Google Scholar]
- 48.Wei Y, Tavis JE, Ganem D. Relationship between viral DNA synthesis and virion envelopment in hepatitis B virus. Journal of Virology. 1997;70:6455–6458. doi: 10.1128/jvi.70.9.6455-6458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perlman D, Hu J. Duck hepatitis B virus virion secretion requires a double-stranded DNA genome. J Virol. 2003;77:2287–2294. doi: 10.1128/JVI.77.3.2287-2294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perlman DH, Berg EA, O'Connor PB, Costello CE, Hu J. Reverse transcription-associated dephosphorylation of hepadnavirus nucleocapsids. Proc Natl Acad Sci U S A. 2005;102:9020–9025. doi: 10.1073/pnas.0502138102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu TT, Coates L, Aldrich CE, Summers J, Mason WS. In hepatocytes infected with duck hepatitis B virus, the template for viral RNA synthesis is amplified by an intracellular pathway. Virology. 1990;175:255–261. doi: 10.1016/0042-6822(90)90206-7. [DOI] [PubMed] [Google Scholar]
- 52.Lenhoff RJ, Summers J. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J Virol. 1994;68:4565–4571. doi: 10.1128/jvi.68.7.4565-4571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Summers J, Smith PM, Horwich AL. Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. Journal of Virology. 1990;64:2819–2824. doi: 10.1128/jvi.64.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Block TM, Mehta AS, Blumberg BS, Dwek RA. Does rapid oligomerization of hepatitis B envelope proteins play a role in resistance to proteasome degradation and enhance chronicity? DNA & Cell Biology. 2006;25:165–170. doi: 10.1089/dna.2006.25.165. [DOI] [PubMed] [Google Scholar]
- 55.Ostapchuk P, Hearing P, Ganem D. A dramatic shift in the transmembrane topology of a viral envelope glycoprotein accompanies hepatitis B viral morphogenesis. EMBO Journal. 1994;13:1048–1057. doi: 10.1002/j.1460-2075.1994.tb06353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bruss V, Ganem D. The role of envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci U S A. 1991;88:1059–1063. doi: 10.1073/pnas.88.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo JT, Pugh JC. Topology of the large envelope protein of duck hepatitis B virus suggests a mechanism for membrane translocation during particle morphogenesis. J Virol. 1997;71:1107–1114. doi: 10.1128/jvi.71.2.1107-1114.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prange R, Streeck RE. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO Journal. 1995;14:247–256. doi: 10.1002/j.1460-2075.1995.tb06998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Persing DH, Varmus HE, Ganem D. The preS1 protein of hepatitis B virus is acylated at its amino terminus with myristic acid. J Virol. 1987;61:1672–1677. doi: 10.1128/jvi.61.5.1672-1677.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu M, Summers J. A domain of the hepadnavirus capsid protein is specifically required for DNA maturation and virus assembly. Journal of Virology. 1991;65:2511–2517. doi: 10.1128/jvi.65.5.2511-2517.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Evans AA, O'Connell AP, Pugh JC, Mason WS, Shen FM, Chen GC, Lin WY, Dia A, M'Boup S, Drame B, London WT. Geographic variation in viral load among hepatitis B carriers with differing risks of hepatocellular carcinoma. Cancer Epidemiology Biomarkers Prevention. 1998;7:559–565. [PubMed] [Google Scholar]
- 62.Chisari FV. Rous-Whipple Award Lecture. Viruses, immunity, and cancer: lessons from hepatitis B. American Journal of Pathology. 2000;156:1117–1132. doi: 10.1016/s0002-9440(10)64980-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rehermann B, Lau D, Hoofnagle JH, Chisari FV. Cytotoxic T lymphocyte responsiveness after resolution of chronic hepatitis B virus infection. Journal of Clinical Investigation. 1996;97:1655–1665. doi: 10.1172/JCI118592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guidotti LG, Morris AG, Mendez H, Koch R, Silverman RH, Williams BR, Chisari FV. Interferon-regulated pathways that control hepatitis B virus replication in transgenic mice. Journal of Virology. 2002;76:2617–2621. doi: 10.1128/JVI.76.6.2617-2621.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Webster JMG, Reignat S, Maini MK, Whalley SA, Ogg GS, King A, Brown D, Amlot PL, Williams R, Vergani D, Dusheiko GM, Bertoletti A. Incubation phase of acute hepatitis B in man: dynamic of cellular immune mechanisms. Hepatology. 2000;32:1117–1124. doi: 10.1053/jhep.2000.19324. [DOI] [PubMed] [Google Scholar]
- 66.Webster G, Bertoletti A. Quantity and quality of virus-specific CD8 cell response: relevance to the design of a therapeutic vaccine for chronic HBV infection. Molecular Immunology. 2001;38:467–473. doi: 10.1016/s0161-5890(01)00082-7. [DOI] [PubMed] [Google Scholar]
- 67.Su AI, Pezacki JP, Wodicka L, Brideau AD, Supekova L, Thimme R, Wieland S, Bukh J, Purcell RH, Schultz PG, Chisari FV. Genomic analysis of the host response to hepatitis C virus infection. Proc Natl Acad Sci U S A. 2002;99:15669–15674. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wieland SF, Guidotti LG, Chisari FV. Intrahepatic induction of alpha/beta interferon eliminates viral RNA-containing capsids in hepatitis B virus transgenic mice. J Virol. 2000;74:4165–4173. doi: 10.1128/jvi.74.9.4165-4173.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Geissler M, Bruss V, Michalak S, Hockenjos B, Ortmann D, Offensperger WB, Wands JR, Blum HE. Intracellular retention of hepatitis B virus surface proteins reduces interleukin-2 augmentation after genetic immunizations. J Virol. 1999;73:4284–4292. doi: 10.1128/jvi.73.5.4284-4292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu Y, Simsek E, Norton P, Sinnathamby G, Philip R, Block TM, Zhou T. The role of the downstream signal sequences in the maturation of the HBV Middle Surface glycoprotein: Development of a novel therapeutic vaccine candidate. Virology. 2007 doi: 10.1016/j.virol.2007.03.042. In press. [DOI] [PubMed] [Google Scholar]
- 71.Gozuacik D, Murakami Y, Saigo K, Chami M, Mugnier C, Lagorce D, Okanoue T, Urashima T, Brechot C, Paterlini-Brechot P. Identification of human cancer-related genes by naturally occurring Hepatitis B Virus DNA tagging. Oncogene. 2001;20:6233–6240. doi: 10.1038/sj.onc.1204835. [DOI] [PubMed] [Google Scholar]
- 72.Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. Journal of Virology. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dragani TA, Manenti G, Farza H, Della Porta G, Tiollais P, Pourcel C. Transgenic mice containing hepatitis B virus sequences are more susceptible to carcinogen-induced hepatocarcinogenesis. Carcinogenesis. 1990;11:953–956. doi: 10.1093/carcin/11.6.953. [DOI] [PubMed] [Google Scholar]
- 74.Zhou YZ, Slagle BL, Donehower LA, vanTuinen P, Ledbetter DH, Butel JS. Structural analysis of a hepatitis B virus genome integrated into chromosome 17p of a human hepatocellular carcinoma. Journal of Virology. 1998;62:4224–4231. doi: 10.1128/jvi.62.11.4224-4231.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feitelson MA. Hepatitis B virus in hepatocarcinogenesis. Journal of Cellular Physiology. 1999;181:188–202. doi: 10.1002/(SICI)1097-4652(199911)181:2<188::AID-JCP2>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 76.Benn J, Schneider RJ. Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signaling cascade. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:10350–10354. doi: 10.1073/pnas.91.22.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang XW, Forrester K, Yeh H, Feitelson MA, Gu JR, Harris CC. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc Natl Acad Sci USA. 1994;91:2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee YI, Kang-Park S, Do SI, Lee YI. The hepatitis B virus-X protein activates a phosphatidylinositol 3-kinase-dependent survival signaling cascade. J Biol Chem. 2001;276:16969–16977. doi: 10.1074/jbc.M011263200. [DOI] [PubMed] [Google Scholar]
- 79.Chisari F, Klopchin K, Morijama T, Pasquinelli C, Dunsford HA, Sell S, Pinkert CA, Brinster RL, Palmiter RD. Molecular pathogenesis of hepatocellular carcinoma in hepatitis B virus transgenic mice. Cell. 1989;59:1145–1156. doi: 10.1016/0092-8674(89)90770-8. [DOI] [PubMed] [Google Scholar]
- 80.Hildt E, Urban S, Hofschneider PH. Characterization of essential domains for the functionality of the MHBst transcriptional activator and identification of a minimal MHBst activator. Oncogene. 1995;11:2055–2066. [PubMed] [Google Scholar]
- 81.Hildt E, Urban S, Lauer U, Hofschneider PH, Kekule AS. ER-localization and functional expression of the HBV transactivator MHBst. Oncogene. 1993;8:3359–3367. [PubMed] [Google Scholar]
- 82.Hildt E, Munz B, Saher G, Reifenberg K, Hofschneider PH. The PreS2 activator MHBs(t) of hepatitis B virus activates c-raf-1/Erk2 signaling in transgenic mice. Embo J. 2002;21:525–535. doi: 10.1093/emboj/21.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ciupe SM, Ribeiro RM, Nelson PW, Dusheiko G, Perelson AS. The role of cells refractory to productive infection in acute hepatitis B viral dynamics. Proceedings of the National Academy of Sciences. 2007;104:5050–5055. doi: 10.1073/pnas.0603626104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang P, Yu MY, Venable R, Alter HJ, Shih JW. Neutralization epitope responsible for the hepatitis B virus subtype-specific protection in chimpanzees. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:9214–9219. doi: 10.1073/pnas.0603316103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu ZY, Liu CB, Francis DP, Purcell RH, Gun ZL, Duan SC, Chen RJ, Margolis HS, Huang CH, Maynard JE. Prevention of perinatal acquisition of hepatitis B virus carriage using vaccine: preliminary report of a randomized, double-blind placebo-controlled and comparative trial. Pediatrics. 1985;76:713–718. [PubMed] [Google Scholar]
- 86.CDC. General recommendations on immunization: Recommendations of the Advisory Committee on Immunization Practices (ACIP) and the American Academy of Family Physicians (AAFP) MMWR: Morbidity and Mortality Weekly Report. 2002;51:1–36. [PubMed] [Google Scholar]
- 87.CDC. Immunization of Health-Care Workers: Recommendations of the Advisory Committee on Immunization Practices (ACIP) and the Hospital Infection Control Practices Advisory Committee (HICPAC) MMWR: Morbidity and Mortality Weekly Report . 1997;46:1–42. [PubMed] [Google Scholar]
- 88.Paulij WP, de Wit PL, Sunnen CM, van Roosmalen MH, Petersen-van Ettekoven A, Cooreman MP, Heijtink RA. Localization of a unique hepatitis B virus epitope sheds new light on the structure of hepatitis B virus surface antigen. Journal of General Virology. 1999;80:2121–2126. doi: 10.1099/0022-1317-80-8-2121. [DOI] [PubMed] [Google Scholar]
- 89.Zuckerman JN, Zuckerman AJ. Current topics in hepatitis B. Journal of Infection. 2000;41:130–136. doi: 10.1053/jinf.2000.0720. [DOI] [PubMed] [Google Scholar]
- 90.Schochetman G, Kuhns MC. HIV variants and hepatitis B surface antigen mutants: diagnostic challenges for immunoassays. Journal of Medical Virology. 2006;78:S3–S6. doi: 10.1002/jmv.20598. [DOI] [PubMed] [Google Scholar]
- 91.Wieland SF, Eustaquio A, Whitten-Bauer C, Boyd B, Chisari FV. Interferon prevents formation of replication-competent hepatitis B virus RNA-containing nucleocapsids. Proc Natl Acad Sci U S A. 2005;102:9913–9917. doi: 10.1073/pnas.0504273102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Davis MG, Jansen RW. Inhibition of hepatitis B virus in tissue culture by alpha interferon. Antimicrobial Agents & Chemotherapy. 1994;38:2921–2924. doi: 10.1128/aac.38.12.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lau GK, Piratvisuth T, Luo KX, Marcellin P, Thongsawat S, Cooksley G, Gane E, Fried MW, Chow WC, Paik SW, Chang WY, Berg T, Flisiak R, McCloud P, Pluck N. Peginterferon Alfa-2a, lamivudine, and the combination for HBeAg-positive chronic hepatitis B. N Engl J Med. 2005;352:2682–2695. doi: 10.1056/NEJMoa043470. [DOI] [PubMed] [Google Scholar]
- 94.Liaw YF, Leung NW, Chang TT, Guan R, Tai DI, Ng KY, Chien RN, Dent J, Roman L, Edmundson S, Lai CL. Effects of extended lamivudine therapy in Asian patients with chronic hepatitis B. Asia Hepatitis Lamivudine Study Group. Gastroenterology. 2000;119:172–180. doi: 10.1053/gast.2000.8559. [DOI] [PubMed] [Google Scholar]
- 95.Torresi J. The virological and clinical significance of mutations in the overlapping envelope and polymerase genes of hepatitis B virus. Journal of Clinical Virology. 2002;25:97–106. doi: 10.1016/s1386-6532(02)00049-5. [DOI] [PubMed] [Google Scholar]
- 96.King RW, Ladner SK, Miller TJ, Zaifert K, Perni RB, Conway SC, Otto MJ. Inhibition of human hepatitis B virus replication by AT-61, a phenylpropenamide derivative, alone and in combination with (-)beta-L- 2',3'-dideoxy-3'-thiacytidine. Antimicrob Agents Chemother. 1998;42:3179–3186. doi: 10.1128/aac.42.12.3179. [published erratum appears in Antimicrob Agents Chemother 1999 Mar;43(3):726] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Deres K, Schroder CH, Paessens A, Goldmann S, Hacker HJ, Weber O, Kramer T, Niewohner U, Pleiss U, Stoltefuss J, Graef E, Koletzki D, Masantschek RN, Reimann A, Jaeger R, Gross R, Beckermann B, Schlemmer KH, Haebich D, Rubsamen-Waigmann H. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science. 2003;299:893–896. doi: 10.1126/science.1077215. [DOI] [PubMed] [Google Scholar]
- 98.Weber O, Schlemmer KH, Hartmann E, Hagelschuer I, Paessens A, Graef E, Deres K, Goldmann S, Niewoehner U, Stoltefuss J, Haebich D, Ruebsamen-Waigmann H, Wohlfeil S. Inhibition of human hepatitis B virus (HBV) by a novel non-nucleosidic compound in a transgenic mouse model. Antiviral Res. 2002;54:69–78. doi: 10.1016/s0166-3542(01)00216-9. [DOI] [PubMed] [Google Scholar]