Abstract
All eukaryotic cells analyzed have developed mechanisms to eliminate the production of mRNAs that prematurely terminate translation. The mechanisms are thought to exist to protect cells from the deleterious effects of in-frame nonsense codons that are generated by routine inefficiencies and inaccuracies in RNA metabolism such as pre-mRNA splicing. Depending on the particular mRNA and how it is produced, nonsense codons can mediate a reduction in mRNA abundance either (i) before its release from an association with nuclei into the cytoplasm, presumably but not certainly while the mRNA is being exported to the cytoplasm and translated by cytoplasmic ribosomes, or (ii) in the cytoplasm. Here, we provide evidence for a factor that functions to eliminate the production of nonsense-containing RNAs in mammalian cells. The factor, variously referred to as Rent1 (regulator of nonsense transcripts) or HUPF1 (human Upf1 protein), was identified by isolating cDNA for a human homologue to Saccharomyces cerevisiae Upf1p, which is a group I RNA helicase that functions in the nonsense-mediated decay of mRNA in yeast. Using monkey COS cells and human HeLa cells, we demonstrate that expression of human Upf1 protein harboring an arginine-to-cysteine mutation at residue 844 within the RNA helicase domain acts in a dominant-negative fashion to abrogate the decay of nonsense-containing mRNA that takes place (i) in association with nuclei or (ii) in the cytoplasm. These findings provide evidence that nonsense-mediated mRNA decay is related mechanistically in yeast and in mammalian cells, regardless of the cellular site of decay.
All eukaryotic cells are thought to be characterized by pathways that eliminate the production of mRNAs that encode truncated proteins (reviewed in refs. 1–4). Truncated proteins can arise as the consequence of inefficiencies or inaccuracies in RNA metabolism. In fact, errors in transcription initiation and splicing can result in the production of either nonfunctional protein or protein that acts in a dominant-negative or gain-of-function fashion and may have provided the selective pressure under which the nonsense-mediated decay pathway evolved (reviewed in ref. 2). The nonsense-mediated decay pathway also eliminates nonsense-containing mRNAs for Igs and T cell receptors that arise as a consequence of nonproductive gene rearrangements and hypermutations (reviewed in ref. 5), certain selenoprotein mRNAs when a UGA codon is recognized as a premature termination codon rather than a selenocysteine codon (6), and nonsense-containing mRNAs that arise as the consequence of random germ-line or somatic defects (reviewed in ref. 2).
Trans-acting factors required for nonsense-mediated mRNA decay in S. cerevisiae have been defined by genetic analyses to include Upf1p/Isf2p/Sal2p/Nam7p, Upf2p/Nmd2p/Sua1p/Ifs1p, Upf3p/Sua6p, Xrn1p, which is a 5′ → 3′ exonuclease, and Dcp1p, which is a decapping enzyme (7–19). Evidence that the three Upf factors function in a common pathway that leads to the accelerated decay of nonsense-containing mRNA derives from the findings that (i) strains doubly null for pairwise combinations of the three UPF genes or triply null for all three UPF genes have nonadditive effects on the decay of nonsense-containing mRNAs (8, 13, 15–17, 20, 21), (ii) physical interactions between Upf1p and Upf2p and between Upf2p and Upf3p have been detected by using two-hybrid analyses (15), and (iii) all three factors colocalize with 80S ribosome particles and polyribosomes (11, 20, 22). Upf1p, the Upf factor best characterized biochemically, demonstrates RNA/DNA-dependent ATPase activity, 5′ → 3′ helicase activity, and can bind to DNA or RNA better in the absence of ATP than in the presence of ATP (23, 24). In addition to its role in nonsense-mediated mRNA decay, Upf1p is thought to function in translation termination. A UPF1Δ strain of Saccharomyces cerevisiae was shown to have a nonsense suppression phenotype, and mutations within the UPF1 gene demonstrated distinct regions that modulate Upf1p function in nonsense mRNA decay and in translation termination at a nonsense codon (24, 25).
The recent isolation of cDNA for the human homologue to yeast Upf1p revealed 51% identity between the human and yeast proteins, and conservation of (i) the cysteine/histidine-rich region having zinc finger-like features, (ii) the seven motifs characteristic of group I RNA helicases, (iii) the distances between six of the seven helicase motifs, and (iv) all amino acids known to be critical for yeast Upf1p function (26, 27). Human (h) Upf1 protein (p), which consists of 1,118 aa, differs from yeast Upf1p, which consists of 853 aa, most noticeably by having 63 additional amino acids at the N-terminal end and 83 additional amino acids at the C-terminal end. The additional N-terminal end is composed of an acidic stretch of 34 aa followed by three tandem repeats of a proline/glycine-rich sequence, whereas the additional C-terminal end consists of several SQ and SQP repeats of unknown significance. Remarkably, analysis of part of the mouse UPF1 gene revealed that amino acids 78 through 1,118 of mouse Upf1p differed from hUpf1p at only 14 residues (26). Western blot analysis of extracts from human and mouse cells using an anti-hUpf1 peptide antibody revealed a single band with an apparent molecular mass of 130 kDa, consistent with the size of product synthesized in vitro from hUPF1 cDNA (27). In agreement with localization of yeast Upf1p to the cytoplasm, hUpf1p was detected exclusively in the cytoplasm by immunofluorescence staining of ethanol-fixed human and mouse cells and exclusively in postnuclear fractions by Western blot hybridization of extracts from human and mouse cells (27). However, unlike yeast Upf1p, hUpf1p is characterized by a sequence KKLK(X17)KKR that is similar to the consensus for a bipartite nuclear localization signal (26). The only functional assay for hUpf1p to date has been the demonstration that expression of a chimeric protein, containing the central region of hUpf1p flanked by the extreme N and C termini of yeast Upf1p, complements Upf1p-deficient yeast in a frameshift allosuppression assay, indicating function in translation termination (26).
To determine whether hUpf1p functions in the nonsense-mediated decay of mRNA in mammalian cells, hUPF1 cDNA, either with or without an arginine-to-cysteine mutation at residue 844 (R844C), was inserted into the plasmid pCI-neo and transiently introduced into monkey COS cells. The R844C mutation of hUPF1 cDNA corresponds to the so-called D4 mutation of the yeast UPF1 gene (8). This mutation converts the conserved arginine at residue 779 within the RNA helicase domain to a cysteine and has been shown to confer a dominant-negative inhibition of yeast Upf1p effect on nonsense-mediated mRNA decay (8) while retaining the ability to associate with polyribosomes (20). We reasoned that if hUpf1p were to function in mammalian cells as it does in yeast, then expression of R844C hUPF1 cDNA in COS cells might eliminate decay in a dominant-negative fashion. Results indicate that this is, indeed, the case: expression of R884C hUPF1 cDNA abrogates the nonsense-mediated decay of β-globin (Gl) mRNA and selenium-dependent glutathione peroxidase 1 (GPx1) mRNA. Notably, nonsense-containing Gl mRNA is degraded in association with nuclei (28, 29), whereas nonsense-containing GPx1 mRNA is degraded in the cytoplasm (6). hUPF1 cDNA harboring the R844C mutation also was inserted into pFlag-IRES1neo and stably introduced into human HeLa cells. As with COS cells that transiently express R844C hUPF1 cDNA, HeLa cells that stably express R844C hUPF1 cDNA also abrogate in the decay of nonsense-containing mRNA. These data provide evidence for a factor that functions in the nonsense-mediated decay of mRNA in mammalian cells and indicate that nonsense-mediated mRNA decay is mechanistically related in yeast and mammals, regardless of the cellular site of decay in mammals.
METHODS
Plasmid Constructions.
To construct pCI-neo-hUPF1, pCMVSport-Rent1 (1) was digested with XhoI and BlpI and treated with Klenow and dNTPs. The resulting 3.38-kbp XhoI-BlpI fragment was inserted into pCI-neo (Promega) that had been digested with XhoI and XbaI and treated with Klenow and dNTPs. In so doing, hUPF1 cDNA was preceded by the human cytomegalovirus (CMV) promoter, a 145-bp exon and a 132-bp chimeric β-globin-IgG intron, and followed by the simian virus 40 (SV40) late polyadenylation sequence. pCI-neo-hUPF1 R844C, in which the arginine codon (CGG) for hUpf1p residue 844 is converted to a cysteine codon (TGT), was constructed by ligating the 1.08-kbp AflIII-PmlI fragment from pβUPF1 R844C to the 7.81-kbp PmlI-AflIII fragment of pCI-neo-hUPF1. The R844C mutation was generated by using the Altered Sites II in vitro mutagenesis kit (Promega) and the antisense mutagenic oligonucleotide 5′-GTCCTGTGTGTGTGCCAAC-3′.
pFlag-hUPF1-IRES1neo was constructed by inserting a Klenow-treated, 4.3-kbp NheI-NotI fragment containing the Flag sequence in addition to hUPF1 cDNA from pFlag-hUPF1-neo harboring the R844C mutation into the EcoRV and NotI sites within the polylinker region of pIRESneo (CLONTECH). To construct pFlag-hUPF-neo, the 1.92-kbp XhoI-MscI fragment from pCMVSport-Rent1 was inserted into the XhoI and EcoRV sites of pBluescript KS(−) after Klenow-filling the MscI site. A HindIII site was generated at the hUPF1 initiation codon to destroy the codon by using the mutagenic sense oligonucleotide 5′-GAGGCAAGCTTAGCGTGGAGG-3′ (nucleotides that comprise the HindIII site are underlined). The 1.78-kbp HindIII-KpnI fragment from the pBluescript subclone that includes UPF1 cDNA then was ligated to the 1.33-kbp KpnI-BamHI fragment from pCI-neo, which includes the SV40 enhancer–early promoter and the neo gene, and the 3.98-kbp BamHI-HindIII fragment from pFlag-CMV-2 (Kodak), which includes the Escherichia coli β-lactamase gene and origin of replication. Before purifying the pFlag-CMV-2 fragment, the hGH polyadenylation sequence and most of the origin of SV40 replication (1.14 kbp) were removed from the plasmid by ligating the SmaI site within the polylinker to the Klenow-filled AvrII site within the SV40 origin. A Klenow-filled, 335-bp HindIII-XhoI fragment from pCI-neo that harbors the hybrid β-globin-IgG intron was inserted into the Klenow-filled NcoI site that overlaps the Flag initiation codon. Insertion was made after subcloning the 55-bp SacI-HindIII fragment, which harbors the NcoI site, into pUC19. pFlag-hUPF1-neo constructs were generated in two steps: (i) ligating the 1.78-kbp HindIII-KpnI fragment, the 1.33-kbp KpnI-BamHI fragment that extends from the SV40 enhancer–promoter region to beyond the poly(A) site of the neo gene, and the 3.98-kbp BamHI-HindIII fragment that extends from beyond the poly(A) site to the polylinker region of pFlag-CMV-2 and harbors the inserted β-globin-IgG intron and (ii) inserting the missing hUPF1 cDNA sequences that harbor the R844C mutation plus sequences up to and including the SV40 promoter–enhancer as a 2.54-kbp KpnI-KpnI fragment into the sole KpnI site of hUPF1 cDNA.
Cell Transfections, Analysis of Transfection Efficiencies, and RNA Purification.
Monkey kidney COS cells were propagated in minimal essential medium containing 10% fetal calf serum and 5% bovine calf serum and transiently transfected by using calcium phosphate (29) and a total of 50 μg of plasmid DNA. To determine transfection efficiencies, cells were transfected with pEGFP-N1 (CLONTECH), fixed with 4% formaldehyde in PBS 48 hr after transfection, counter-stained with 4′,6-diamidino-2-phenylindole, and visualized by using a Nikon Optiphot-2 fluorescence microscope. HeLa cells (at 30% confluency in a 15-cm diameter plate) were propagated in minimal essential medium containing 10% fetal calf serum and 5% bovine calf serum and stably transfected with 50 μg of pFlag-hUPF1-IRES1neo harboring the R844C mutation using calcium phosphate (29). Fresh medium was added 16 hr after transfection, and, after an additional 24 hr, cells were diluted 15-fold and replated and cultured in the presence of 1 mg/ml of G418 (GIBCO/BRL). Single G418-resistant colonies were isolated and expanded after 10–15 days in selective medium. HeLa cells were transiently transfected by using calcium phosphate and a total of 43 μg of plasmid DNA. Total RNA was purified by using Trizol (GIBCO/BRL). Nuclear and cytoplasmic RNAs were isolated using citric acid and purified using cesium chloride centrifugation (29).
Northern Blot Hybridizations.
To detect Gl and mouse major urinary protein (MUP) RNAs, nuclear or cytoplasmic RNA (40 μg) was denatured, electrophoresed in a 1.5% agarose gel, transferred to a nylon membrane, and hybridized to two DNA fragments that had been 32P-labeled by random priming (30, 31).
Antibody Generation, Protein Purification, and Western Blotting.
Rabbit polyclonal antiserum was raised (Covance, Denver, PA) against GST-hUpf1p that included the first 266 aa of hUpf1p. hUpf1p antibody was then purified by binding in 10 mM Na2HPO4 (pH 7.0) and 0.5M NaCl to GST-hUpf1p conjugated to CNBr-activated Sepharose 4B (Pharmacia), elution in 0.1 M glycine-HCl (pH 2.5), neutralization in 1 M Hepes-KOH (pH 7.4), and dialysis in PBS. Total-cell protein was isolated from untransfected or transfected cells by boiling in 0.1 M Tris⋅HCl, pH 6.8/10% glycerol/2% SDS/2-mercaptoethanol. Serial dilutions of 3 or 30 μg from transfected cells and 30 μg from untransfected cells were electrophoresed in a 7.5% polyacrylamide gel, transferred to nitrocellulose, probed with polyclonal anti-hUpf1p and a secondary peroxidase-conjugated anti-rabbit IgG (Amersham), and visualized by using enhanced chemiluminescence (ECL; Amersham).
Reverse Transcription–PCR (RT-PCR) Analyses.
For RT-PCR, 10 μg of total, nuclear, or cytoplasmic RNA was treated with 1 unit of RNase-free RQ DNase I (Promega), cDNA was synthesized from 0.33–3.0 μg of the RNA using Moloney murine leukemia virus reverse transcriptase (Superscript II; GIBCO/BRL), and PCR was used to amplify the resulting cDNA (30, 32, 33). Primers for the amplification of hUPF1 RNA consisted of 5′-CCTGCTGCAGGGCGAGGCAC-3′ (sense, corresponding to nucleotides 3007–3026, where nucleotide 1 is defined as the first nucleotide of the hUPF1 initiation codon), and 5′-CTGCATTCTAGTTGTGGTTTG-3′ (antisense, corresponding to the SV40 polyadenylation region). Primers for the simultaneous amplification of hUPF1 and COS cell UPF1 RNAs consisted of 5′-GCCAGCGCTCCTACCTG-3′ (sense, corresponding to nucleotides 2393–2403 of hUPF1 RNA) and 5′-GCCACGTTCAGACGCC-3′ (antisense, corresponding to nucleotides 2585–2570 of hUPF1 RNA). Primers for the amplification of MUP RNA from exon 4 into exon 7 (31), GPx1 RNA from the mCMV 5′ untranslated region into exon 2 (6, 34), and Gl RNA from the human 5′ untranslated region (32) into mouse exon (29) have been described.
RESULTS
Transient Expression of R844C hUPF1 cDNA in COS Cells Abrogates the Nucleus-Associated Decay of Nonsense-Containing β-Globin mRNA.
With the aim of determining whether hUpf1p functions in the nonsense-mediated decay of mRNA in mammalian cells, hUPF1 cDNA that does or does not harbor an arginine-to-cysteine mutation at residue 844 (R844C) was inserted into the plasmid pCI-neo and transiently introduced into monkey COS cells together with a test plasmid and a reference plasmid. The R844C mutation was chosen for analysis because the corresponding R779C mutation within the yeast UPF1 gene has been shown to confer the strongest dominant-negative inhibition of yeast Upf1p function of all dominant-negative Upf1p mutations characterized to date (8). A 3.3-fold accumulation of nonsense-containing his4–38 mRNA was detected when a yeast UPF1 gene carrying the R779C (or D4) mutation was placed in a multicopy plasmid and expressed in a yeast strain carrying a wild-type UPF1 gene (8). This is an appreciable fraction of the 4.4-fold accumulation detected for his4–38 mRNA when the UPF1 gene carrying the R779C mutation was expressed in yeast carrying a deletion within the UPF1 gene that completely eliminates Upf1p function (7). Expression of either wild-type or mutated hUPF1 cDNA in COS cells was augmented by (i) the SV40 origin of replication within pCI-neo, which allows for transient episomal replication of the cDNA in the presence of the COS cell-encoded SV40 large T antigen, (ii) efficient transcription of hUPF1 cDNA directed by the strong human CMV (hCMV) promoter, and (iii) efficient production of hUPF1 mRNA because of an efficiently spliced intron positioned between the transcription start site and hUPF1 cDNA.
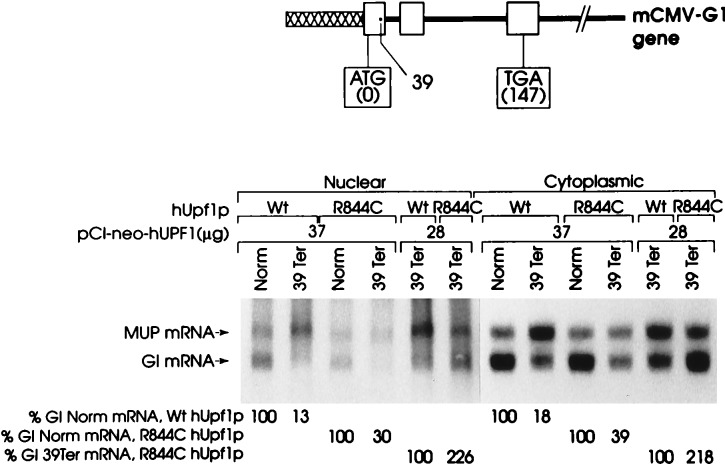
The test plasmid, pmCMV-Gl, harbored a human–mouse hybrid Gl gene that was either nonsense-free or contained a nonsense mutation within codon 39 of exon 1 and was driven by the mouse CMV (mCMV) promoter (ref. 29; Fig. 1). The reference plasmid, phCMV-MUP (31), harbored the MUP gene driven by the human CMV promoter and served to control for variations in the efficiencies of cell transfection and RNA recovery. Cells were harvested 48 hr after transfection, and the amounts of Gl and MUP RNAs in nuclear and cytoplasmic fractions were quantitated by using Northern blot hybridization. For each transfection, the amount of Gl mRNA was normalized to the amount of MUP mRNA and presented as a percentage of the normalized amount of either nonsense-free (Norm) or nonsense-containing (39Ter) Gl mRNA.
Figure 1.
Transient expression of R(844)C hUPF1 cDNA in COS cells abrogates the nonsense-mediated decrease in the abundance of β-globin mRNA. COS cells were transiently transfected with a pmCMV-Gl test plasmid (either Norm, which is nonsense-free, or 39Ter, which harbors a nonsense codon at amino acid position 39), the phCMV-MUP reference plasmid, and pCI-neo-hUPF1 [either Wt, which harbors an unmutagenized hUpf1p reading frame, or R(844)C, which harbors the arginine-to-cysteine change at amino acid position 844]. The amounts of each plasmid used were, respectively, 10 μg, 3 μg, and 37 μg, or 19 μg, 3 μg, and 28 μg. An appropriate amount of a fourth plasmid was added to each transfection to bring the total amount of introduced DNA to 50 μg. Nuclear and cytoplasmic RNA was purified (29, 30), and 40 μg were analyzed by blot hybridization to detect globin (Gl) RNA and mouse major urinary protein (MUP) RNA. Hybridization was quantitated by PhosphorImaging. The level of Gl mRNA from each mCMV-Gl allele was normalized to the level of MUP mRNA to provide a quantitative analysis. Normalized values then were calculated as a percentage of the normalized value of Gl Norm mRNA in the presence of either the Wt hUPF1 gene or the R844C hUPF1 gene or as a percentage of the normalized value of Gl 39Ter mRNA in the presence of the R844C hUPF1 gene, each of which was considered as 100. Percentages differed between two independently performed experiment by no more than 7%.
Transfecting cells with 37 μg of pCI-neo-hUPF1 that harbors wild-type (Wt) hUPF1 cDNA did not preclude the 39Ter-mediated decay of Gl mRNA (29), because the level of 39Ter mRNA was 13% and 18% of normal for nuclear and cytoplasmic fractions, respectively (Fig. 1). In contrast, transfecting cells with 37 μg of pCI-neo-hUPF1 that harbors hUPF1 cDNA containing the R844C mutation abrogated the 39Ter-mediated decay of Gl mRNA so the level of 39Ter mRNA was 30% and 39% of normal for nuclear and cytoplasmic fractions, respectively (Fig. 1). This 2.2- to 2.3-fold increase in the level of 39Ter mRNA is comparable to the increase mediated when 28 μg of pCI-neo-hUPF1 containing the R844C mutation was used (Fig. 1). Therefore, analogous to overproducing Upf1p harboring the R779C mutation in S. cerevisiae (8, 20), overproducing hUpf1p harboring the R844C mutation in mammalian cells partially restores the level of nonsense-containing mRNA to normal and implicates a role for hUpf1p in nonsense-mediated decay in mammalian cells.
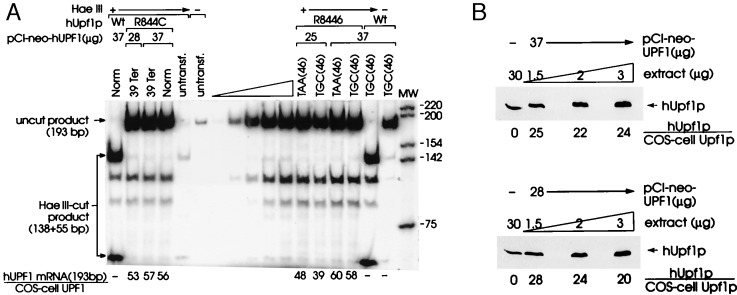
To better understand the basis of the partial abrogation of nonsense-mediated decay, the level of exogenous hUPF1 RNA was compared with the level of COS cell UPF1 RNA by using RT-PCR and a primer pair that amplifies both RNAs and generates a 193-bp product. The RT-PCR products of each RNA could be distinguished in transfections involving R844C hUPF1 cDNA because the R844C mutation destroys a HaeIII site: whereas the 193-bp product that derives from R844C hUPF1 RNA is resistant to cleavage by HaeIII, the 193-bp product that derives from COS cell UPF1 RNA is cleaved by HaeIII to 138-bp and 55-bp fragments. HaeIII digestions were demonstrated to be complete by spiking the RT-PCRs with 1 μg of pUC19 DNA and then visualizing the complete cleavage of pUC19 DNA after electrophoresis in agarose and staining with ethidium bromide (data not shown).
The analysis of cytoplasmic RNA indicates that the ratio of R884C hUPF1 RNA to COS cell UPF1 RNA was approximately 57:1 and 53:1 with the use of 37 μg and 28 μg of pCI-neo-hUPF1, respectively (Fig. 2A). These ratios indicate that there was a vast and comparable excess of hUPF1 RNA relative to COS cell UPF1 RNA with either amount of pCI-neo-hUPF1. Consistent with the amount of hUPF1 RNA being comparable regardless of the amount of pCI-neo-hUPF1 used, the ratio of hUPF1 RNA to MUP RNA was identical with the use of either amount of pCI-neo-hUPF1 (data not shown). Considering that approximately 2% of cells were transfected on the basis of the percentage of cells that fluoresce after transfection with pEGFP-N1, a plasmid that produces green fluorescent protein (data not shown), the ratio of hUPF1 RNA to COS cell RNA in transfected cells can be estimated to be 2,750:1. Consistent with this estimation, the sum of exogenous hUpf1p and COS cell Upf1p in total-cell protein was estimated by Western blotting using affinity-purified rabbit polyclonal antibody to hUpf1p and ECL to be 24-fold above the level of COS cell Upf1p in untransfected cells, regardless of whether 37 μg or 28 μg of p-CI-neo-hUPF1 was used (Fig. 2B). Given that only 2% of cells were transfected, and assuming that the level of COS cell Upf1p was unaffected by the production of hUpf1p, the ratio of hUpf1p to COS cell Upf1p in transfected cells is estimated to be 1,200:1. Therefore, the difference between the amounts of hUPF1 RNA and COS cell UPF1 RNA as well as hUpf1p and COS cell Upf1p in the 2% of cells that were transfected was three orders of magnitude. We conclude that the partial abrogation of 39Ter-mediated mRNA decay is not attributable to an insufficient molar excess of hUpf1p to COS cell Upf1p. Given that the comparably mutated yeast Upf1p-R779C in a yeast strain lacking wild-type Upf1p retains the capability to associate with polysomes but still has 62% of function (22), the partial abrogation evidenced with R884C hUpf1p may reflect failure of the mutation to completely inactivate hUpf1p function, failure of the mutated protein to integrate into or otherwise inactivate all COS cell complexes that are required for decay, or both.
Figure 2.
Analysis of the amounts of hUPF1 RNA and hUpf1 protein in COS cells. (A) Cytoplasmic RNA (2.5 μg) from the same transfections analyzed in Fig. 1 (lanes 1–4) and Fig. 3 (lanes 12–17) was used to quantitate the levels of hUPF1 RNA and COS cell UPF1 RNA (A) and hUPF1 and MUP RNAs (B) by using RT-PCR. (A) The ratio of RT-PCR products from hUPF1 and COS cell UPF1 RNAs for those transfections involving pCI-neo-hUPF1 R844C was calculated after digestion with HaeIII, which does not cleave the 193-bp RT-PCR product of R844C hUPF1 RNA but generates fragments of 138 bp and 55 bp from the 193-bp RT-PCR product of COS-cell UPF1 RNA. HaeIII cleavage of the RT-PCR product of cells transfected with pCI-neo-hUPF1 Wt (lane 1), like HaeIII cleavage of the RT-PCR product of COS cell UPF1 RNA (lane 5), generates 138-bp and 55-bp fragments. The lanes marked with a wedge constitute the analysis of a serial dilution of cytoplasmic RNA and establish that there is a linear relationship between the amount of input RNA and the amount of each RT-PCR product. Indication that HaeIII cleavage was complete derives from a comparison of lanes 5 and 6 and lanes 16 and 17, i.e., all of the 193-bp product that derives from either COS cell UPF1 RNA or Wt hUPF1 RNA was cleaved by HaeIII to 138-bp and 55-bp fragments. (B) Immunoblot analysis of 30 μg of untransfected cell protein and serial dilutions of 3 μg of transfected COS cell protein using affinity-purified rabbit polyclonal anti-hUpf1p, a secondary peroxidase-conjugated anti-rabbit IgG, and ECL. Dilutions were analyzed to establish that there is a linear relationship between the amounts of input protein and immunoreactive Upf1 protein. Total Upf1p is the sum of hUpf1p and COS-cell Upf1p in 30 μg of total-cell protein, where the level of COS cell Upf1p was considered as 1. Notably, hUpf1p and COS cell Upf1p comigrate.
Transient Expression of R884C hUPF1 cDNA in COS Cells Abrogates the Cytoplasmic Decay of Nonsense-Containing GPx1 mRNA.
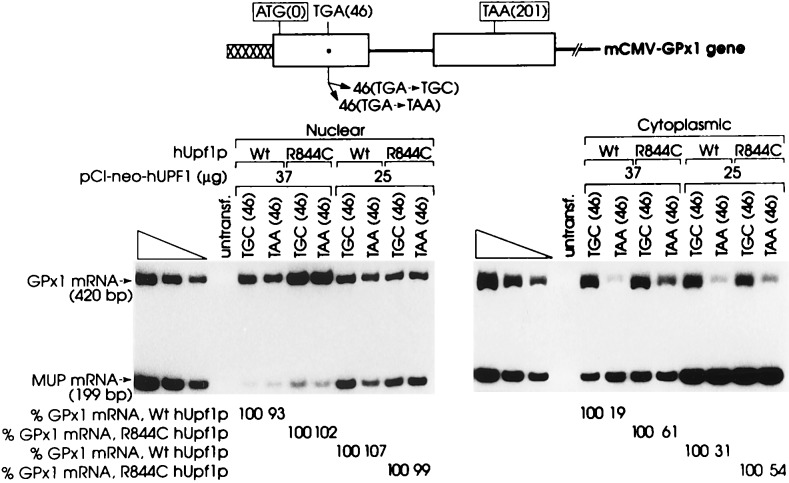
Having demonstrated that hUpf1p functions in the nonsense-mediated decay of Gl mRNA, an mRNA that in non-erythroid cells is subject to nonsense-mediated decay exclusively in association with nuclei (28, 29), probably during the concomitant nuclear export and cytoplasmic translation of the mRNA (1, 29), we sought to determine whether hUpf1p also functions in the nonsense-mediated decay of selenium-dependent glutathione peroxidase 1 (GPx1) mRNA, an mRNA that is subject to nonsense-mediated decay exclusively in the cytoplasm, after being released from an association with nuclei (6). Normally, GPx1 mRNA harbors a UGA codon at amino acid position 46 that inefficiently but purposefully encodes the unusual amino acid selenocysteine under selenium-replete conditions (reviewed in ref. 35). Converting this codon to a UGC cysteine codon eliminates the dependence of gene expression on selenium, allows for efficient translation elongation at position 46 and synthesis of full-length protein terminating at position 201, and obviates nonsense-mediated decay (6). Converting this codon to a UAA nonsense codon also eliminates the dependence of gene expression on selenium but results in efficient translation termination prematurely at position 46 and elicits nonsense-mediated decay even more effectively than does the UGA codon regardless of the selenium concentration (6).
To assess the role of hUpf1p in the nonsense-mediated decay of GPx1 mRNA, COS cells were transiently transfected with pCI-neo-hUPF1 harboring either Wt or R844C hUPF1 cDNA, the phCMV-MUP reference plasmid, and a pmCMV-GPx1 test plasmid in which codon 46 was converted to either a TGC cysteine codon or a TAA nonsense codon. If expression of pCI-neo-hUPF1 that harbors R844C hUPF1 cDNA abrogates the UAA-mediated reduction in the abundance of cytoplasmic GPx1 mRNA, and if expression of pCI-neo-hUPF1 that harbors Wt hUPF1 cDNA is of no consequence to the UAA-mediated reduction in the abundance of cytoplasmic GPx1 mRNA, it can be concluded that hUpf1p functions in cytoplasmic nonsense-mediated mRNA decay.
This was, indeed, the finding. Transfecting cells with either 37 μg or 25 μg of pCI-neo-hUPF1 that harbors Wt hUPF1 cDNA was of no consequence to the relative abundance of GPx1 mRNA produced by the TAA(46)-containing allele or the TGC(46)-containing allele (Fig. 3). The level of UAA(46)-containing GPx1 mRNA was reduced to 19% and 31% of the level of UGC(46)-containing GPx1 mRNA in the cytoplasmic fraction by a mechanism that has no effect on the level of nuclear GPx1 mRNA (Fig. 3). This is consistent with the cytoplasmic level of UAA(46)-containing GPx1 mRNA being reduced to 30% the cytoplasmic level of UGC(46)-containing GPx1 mRNA in the absence of pCI-neo-hUPF1 expression (6). In contrast, transfecting cells with either amount of pCI-neo-hUPF1 that harbors R844C hUPF1 cDNA abrogated the nonsense-mediated reduction of cytoplasmic GPx1 mRNA so that the cytoplasmic level of UAA(46)-containing GPx1 mRNA was 54–61% the level of UGC(46)-containing GPx1 mRNA (Fig. 3). This 1.7- to 3.2-fold increase mediated by R844C hUpf1p is comparable to the level of abrogation evidenced for the nucleus-associated decay of nonsense-containing Gl mRNA (Fig. 1) and indicates that R844C hUpf1p functions comparably at the two cellular sites of decay. As predicted from transfections using pmCMV-Gl (Fig. 1), there was no difference in the extent of abrogation mediated by 37 μg or 25 μg of pCI-neo-hUPF1. The ratio of hUPF1 RNA to COS-cell UPF1 RNA in transfected cells was calculated to be 2,600:1 based on a transfection efficiency of 2% and an average ratio of 109:1 for the two RNAs in the cytoplasmic fraction (Fig. 2A).
Figure 3.
Transient expression of R844C hUPF1 cDNA in COS cells abrogates the nonsense-mediated decrease in the abundance of GPx1 mRNA. COS cells were transiently transfected with a test plasmid pmCMV-GPx (harboring either a TGC cysteine codon or a TAA nonsense codon at position 46), the reference plasmid phCMV-MUP, and pCI-neo-hUPF1 (harboring either Wt or R844C hUPF cDNA). The amounts of each plasmid used were, respectively, 10 μg, 3 μg, and 37 μg or 10 μg, 3 μg, and 25 μg. An appropriate amount of a fourth plasmid was added to each transfection to bring the total amount of introduced DNA to 50 μg. Nuclear and cytoplasmic RNA was purified, and the amounts of GPx1 and MUP mRNAs were quantitated by using RT-PCR. GPx1 and MUP mRNAs generate 420-bp and 199-bp products, respectively. The level of mRNA from each mCMV-GPx1 allele was normalized to the level of MUP mRNA. Normalized values for GPx1 mRNA harboring UAA(46) then were calculated as a percentage of the normalized value of GPx1 mRNA harboring UGC(46) in the presence of either the Wt hUPF1 gene or the R844C hUPF1 gene, each of which was considered as 100. Percentages differed between two independently performed experiments by no more than 9%.
Stable Expression of R844C hUPF1 cDNA in HeLa Cells Abrogates the Nucleus-Associated Decay of Nonsense-Containing β-Globin mRNA.
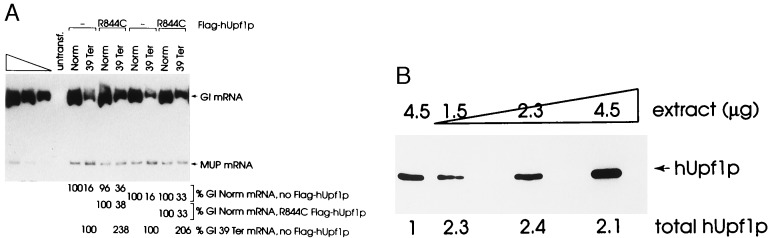
One drawback to elucidating hUpf1p function using data from the transient transfections is the potential for hUpf1p levels that are three orders of magnitude above normal to abrogate nonsense-mediated decay nonspecifically by interfering with an unrelated pathway. Although this possibility is not likely given (i) studies of S. cerevisiae showing that wild-type Upf1p functions in nonsense-mediated decay and Upf1p harboring a mutation comparable to the R844C mutation of hUpfp1 partially abrogates decay (8, 20) and (ii) our studies of COS cells showing that expression of wild-type Upf1p at levels that are three orders of magnitude above normal has no consequence to nonsense-mediated decay (Figs. 1 and 3), we have addressed experimentally the possibility by generating HeLa cells that stably express R844C hUpf1p at a concentration that is closer to normal.
Individual transfectants harboring pFlag-hUPF-IRES1neo R844C were selected by using G418, expanded, and transiently transfected with 25 μg of the pmCMV-Gl test plasmid, either nonsense-free or 39Ter, and 18 μg of the phCMV-MUP reference plasmid. In two separate transfections, R844C hUpf1p increased the level of nonsense-containing Gl mRNA from 16% of normal to 33–38% of normal without significantly affecting the level of nonsense-free Gl mRNA (Fig. 4A). Western blotting using affinity-purified rabbit polyclonal antibody to hUpf1p and ECL demonstrated that the sum of exogenous hUpf1p and HeLa-cell hUpf1p was ≈2.3-fold above the level of hUpf1p in untransfected HeLa cells (Fig. 4B). Together with data obtained from studies of S. cerevisiae, our finding a ≈2-fold increase in the level of nonsense-containing mRNA for cells producing a level of R844C hUpf1p that is either less than an order of magnitude above normal (Fig. 4B) or three orders of magnitude above normal (Fig. 2B) indicates that hUpf1p most likely functions in nonsense-mediated mRNA decay in mammalian cells.
Figure 4.
Stable expression of hUPF1 R(844)C cDNA in HeLa cells abrogates the nonsense-mediated decrease in the abundance of β-globin mRNA. (A) A HeLa cell line stably transfected with pFlag-hUPF1-IRES1neo R844C was transiently transfected with 25 μg of pmCMV-Gl test plasmid (either Norm or 39Ter) and 18 μg of the phCMV-MUP reference plasmid. Total-cell RNA was purified (29, 30), and RT-PCR and PhosphorImaging were used to quantitate Gl and MUP transcripts. Gl and MUP mRNAs generate 486-bp and 199-bp products, respectively. The level of mRNA from each mCMV-Gl allele was normalized to the level of MUP mRNA. Normalized values for Gl mRNA then were calculated as a percentage of the normalized value of Gl Norm mRNA either in the presence (R844C) or absence (−) of the Flag-hUPF1 expression vector, each of which was considered as 100. Percentages differed between two independently performed experiments by no more than 5%. Normalized values for Gl 39Ter mRNA were also calculated as a percentage of the normalized value of Gl 39Ter mRNA in the absence (−) of the Flag-hUPF1 expression vector. (B) Immunoblot analysis of 4.5 μg of protein from untransfected HeLa cells and serial dilutions of 4.5 μg of protein from the HeLa cell line stably transfected with Flag-hUPF1-IRES1neo harboring the R844C mutation. Dilutions were analyzed to establish that there is a linear relationship between the amounts of input protein and immunoreactive hUpf1 protein. Total Upf1p is the sum of R844C hUpf1p and HeLa cell hUpf1p in 4.5 μg of total-cell protein, where the level of hUpf1p was considered as 1.
DISCUSSION
We have obtained evidence using transient and stable cell transfections that the human homologue to S. cerevisiae Upf1p, a factor required for nonsense-mediated mRNA decay in yeast, functions in the nonsense-mediated decay of mRNA in mammalian cells. The success of the experimental approaches taken depended on two conditions. First, success required the production of a sufficient excess of hUpf1p relative to endogenous Upf1p in those cells expressing the test and reference plasmids. Although only a fraction of cells in a transient transfection incorporate the hUPF1 expression vector, the same fraction would be expected to incorporate the test and reference plasmids. Calculations indicate that the transfected fraction produced hUPF1 RNA and hUpf1p in amounts that were three orders of magnitude more than the amounts of endogenous monkey cell UPF1 RNA and Upf1p (Fig. 2). Overexpression of the human RNA and protein to such a large extent was achieved because the hUPF1-containing plasmid is likely to have been introduced at more than one copy per cell, replicates multiple times per cell division, and harbors the relatively strong hCMV promoter to drive hUPF1 gene expression. To achieve a level of overexpression that is closer to the level of endogenous Upf1p, an additional set of experiments was performed by using HeLa cells that were stably transfected with a selectable Upf1p expression vector. Second, success of these experimental approaches required abrogation of nonsense-mediated mRNA decay by the R844C mutation. The finding that the R844C mutation does abrogate nonsense-mediated decay in both the transiently and stably transfected cells (Figs. 1, 3, and 4) is consistent with the possibility that the mutated protein forms a multisubunit complex with one or more endogenous cell proteins so as to inactivate these proteins. Because Upf1p has been shown to enhance translation termination (24, 25, 36), the mutated protein may abrogate nonsense-mediated decay by increasing the level of nonsense suppression. Alternatively, the mutated protein may function more directly in decay, possibly because of decreased RNA binding and/or helicase activities.
Data indicate that hUpf1p functions in the decay of nonsense-containing mRNAs that are degraded either exclusively while nucleus-associated (Figs. 1 and 4) or exclusively while cytoplasmic (Fig. 3). Despite there being two sites of decay in mammalian cells, evidence indicates that Upf1p functions in the cell cytoplasm. First, the biochemical fractionation and in situ immunofluorescence staining of cells demonstrate that the majority of Upf1p in yeast and mammals is cytoplasmic (11, 20, 22, 27). Second, yeast Upf1p, Upf2p, and Upf3p have been shown to interact with polysomes (11, 20, 22). Results of mammalian cell studies that (i) blocked translation using suppressor tRNA, ribosome-binding drugs, a secondary structure within the mRNA 5′ untranslated region, or polio virus infection (31, 37–40) or (ii) allowed for translation reinitiation downstream of a nonsense codon (41) indicate that cytoplasmic ribosomes play a role in nonsense-mediated mRNA decay regardless of whether decay is nucleus-associated or cytoplasmic. Although it is not know how or when Upf1p functions in decay, the finding that yeast Upf1p associates with polysomes but has a concentration that is less than the concentration of ribosomes (20) suggests that some translational process may signal the association of Upf1p with ribosomes. Given the role of yeast Upf1p in translation termination (24, 25, 36), association may take place when a translating ribosome pauses, as it would at a nonsense codon (36).
It is reasonable to hypothesize that the nonsense-mediated decay pathway evolved to eliminate faulty transcripts, such as those arising as a consequence of aberrant gene transcription, ineffective somatic rearrangements of genes encoding Igs and T cell receptors, aberrant or inefficient pre-mRNA splicing, and somatic and inherited mutations. With regard to which transcripts would be subject to nonsense-mediated decay, a survey of genes having one or more 3′ untranslated exons (42), together with studies of several mammalian genes that aimed to distinguish premature termination codons that mediate a reduction in mRNA abundance from premature termination codons that do not (29, 33, 43, 44), have revealed a general rule for mammals and, possibly, other organisms: only those termination codons located more than 50–55 nt upstream of the 3′-most exon–exon junction within mRNA mediate a reduction in mRNA abundance (42). Notably, the 50- to 55-nt measurement applies to fully spliced mRNA, after all introns have been removed by the process of splicing. Exceptions to the rule may include T cell receptor transcripts, in which nonsense codons as close as 8 nt upstream of the 3′-most exon–exon junction mediate a reduction in mRNA abundance (45).
According to the rule, one might expect COS cells overexpressing R844C hUPF1 cDNA to be characterized by an abnormally high cytoplasmic level of pre-mRNAs that fail to be efficiently spliced and harbor intron-derived nonsense codons residing more than 50–55 nt upstream of the 3′-most exon–exon junction. Using RT-PCR, however, there was no detectable cytoplasmic accumulation of either intron 1 of human triosephosphate isomerase pre-mRNA or intron 1 of Gl pre-mRNA (data not shown). These results could indicate that more than a 2- to 3-fold increase in mRNA normally targeted for nonsense-mediated decay would be required to detect accumulation. Alternatively or additionally, each of these introns may be removed sufficiently efficiently so as not to be a significant component of transcripts exported to the cytoplasm. Notably, detection of intron-containing RNA in the cytoplasm of Upf1p-deficient yeast required analysis of an inefficiently removed intron (10, 20, 46). Future experiments are necessary to determine how eliminating nonsense-mediated decay affects cellular metabolism.
Acknowledgments
The authors thank members of the Maquat lab for discussions, Y. Qian and J. Zhang for contributions to plasmid constructions, and J. Bashaw for help with fluorescence microscopy. This work was supported by Public Health Service Research Grants DK33933 and GM52822 (L.E.M.) and GM55239 (H.C.D.) from the National Institutes of Health and the Howard Hughes Medical Institute (H.C.D.).
ABBREVIATIONS
- hUpf1p
human Upf1 protein
- CMV
cytomegalovirus
- SV40
simian virus 40
- Gl
β-globin
- GPx1
selenium-dependent glutathione peroxidase
- MUP
major urinary protein
- RT-PCR
reverse transcription–PCR
- Wt
wild type
References
- 1.Maquat L E. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 2.Maquat L E. Am J Hum Gen. 1996;59:279–286. [PMC free article] [PubMed] [Google Scholar]
- 3.Peltz S W, He F, Welch E, Jacobson A. Prog Nucleic Acid Res Mol Biol. 1994;47:271–298. doi: 10.1016/s0079-6603(08)60254-8. [DOI] [PubMed] [Google Scholar]
- 4.Ruiz-Echevarria M J, Czaplinski K, Peltz S W. Trends Biochem Sci. 1996;21:433–438. doi: 10.1016/s0968-0004(96)10055-4. [DOI] [PubMed] [Google Scholar]
- 5.Alt F W, Blackwell T K, Yancopoulous G D. Science. 1987;238:1079–1087. doi: 10.1126/science.3317825. [DOI] [PubMed] [Google Scholar]
- 6.Moriarty P M, Reddy C C, Maquat L E. Mol Cell Biol. 1998;18:2932–2939. doi: 10.1128/mcb.18.5.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leeds P, Peltz S W, Jacobson A, Culbertson M R. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 8.Leeds P, Wood J M, Lee B S, Culbertson M R. Mol Cell Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altamura N, Groudinsky O, Dujardin G, Slonimski P P. J Mol Biol. 1992;224:575–587. doi: 10.1016/0022-2836(92)90545-u. [DOI] [PubMed] [Google Scholar]
- 10.He F, Peltz S W, Donahue J L, Rosbash M, Jacobson A. Proc Natl Acad Sci USA. 1993;90:7034–7039. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peltz S W, Jacobson A. In: Control of Messenger RNA Stability. Belasco J, Brawerman G, editors. San Diego: Academic; 1993. pp. 291–328. [Google Scholar]
- 12.Muhlrad D, Decker C J, Parker R. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 13.Cui Y, Hagan K W, Zhang S, Peltz S W. Genes Dev. 1995;9:423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 14.Hagan K W, Ruiz-Echevarria M J, Quan Y, Peltz S W. Mol Cell Biol. 1995;15:809–823. doi: 10.1128/mcb.15.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He F, Jacobson A. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- 16.Lee B S, Culbertson M R. Proc Natl Acad Sci USA. 1995;92:10354–10358. doi: 10.1073/pnas.92.22.10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S L, Umen J G, Varmus H E. Proc Natl Acad Sci USA. 1995;92:6587–6591. doi: 10.1073/pnas.92.14.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui Y, Dinman J D, Peltz S W. EMBO J. 1996;15:5726–5736. doi: 10.1002/j.1460-2075.1996.tb00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beelman C A, Stevens A, Caponigro G, LaGrandeur T E, Hatfield L, Fortner D M, Parker R. Nature (London) 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- 20.Atkin A L, Schenkman L R, Eastham M, Dahlseid J N, Lelivelt M J, Culbertson M R. J Biol Chem. 1997;272:22163–22172. doi: 10.1074/jbc.272.35.22163. [DOI] [PubMed] [Google Scholar]
- 21.He F, Brown A H, Jacobson A. Mol Cell Biol. 1997;17:1580–1594. doi: 10.1128/mcb.17.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atkin A L, Altamura N, Leeds P, Culbertson M R. Mol Biol Cell. 1995;6:611–625. doi: 10.1091/mbc.6.5.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Czaplinski K, Weng Y, Hagan Kevin W, Peltz S W. RNA. 1995;1:610–623. [PMC free article] [PubMed] [Google Scholar]
- 24.Weng Y, Czaplinski K, Peltz S W. Mol Cell Biol. 1996;16:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weng Y, Czaplinski K, Peltz S W. Mol Cell Biol. 1996;16:5491–5506. doi: 10.1128/mcb.16.10.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perlick H A, Medghalchi S M, Spencer F A, Kendzior R J, Jr, Dietz H C. Proc Natl Acad Sci USA. 1996;93:10928–10932. doi: 10.1073/pnas.93.20.10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Applequist S E, Selg M, Raman C, Jäck H-M. Nucleic Acids Res. 1997;25:814–821. doi: 10.1093/nar/25.4.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kugler W, Enssle J, Hentze M W, Kulozik A E. Nucleic Acids Res. 1995;23:413–418. doi: 10.1093/nar/23.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Sun X, Qian Y, Maquat L E. RNA. 1998;4:801–815. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng J, Maquat L E. Mol Cell Biol. 1993;13:1892–1902. doi: 10.1128/mcb.13.3.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belgrader P, Maquat L E. Mol Cell Biol. 1994;14:6326–6336. doi: 10.1128/mcb.14.9.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belgrader P, Cheng J, Zhou X, Stephenson L, Maquat L E. Mol Cell Biol. 1994;14:8219–8228. doi: 10.1128/mcb.14.12.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Maquat L E. RNA. 1996;2:235–243. [PMC free article] [PubMed] [Google Scholar]
- 34.Moriarty P M, Reddy C C, Maquat L E. RNA. 1997;3:1369–1373. [PMC free article] [PubMed] [Google Scholar]
- 35.Low S C, Berry M J. Trends Biol Sci. 1996;21:203–208. [PubMed] [Google Scholar]
- 36.Weng Y, Czaplinski K, Peltz S W. RNA. 1998;4:205–214. [PMC free article] [PubMed] [Google Scholar]
- 37.Qian L, Theodor L, Carter M, Vu M N, Sasaki A W, Wilkinson M F. Mol Cell Biol. 1993;13:1686–1696. doi: 10.1128/mcb.13.3.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menon K P, Neufeld E F. Cell Mol Biol. 1994;40:999–1005. [PubMed] [Google Scholar]
- 39.Carter M S, Doskow J, Morris P, Li S, Nhim R P, Sandstedt S, Wilkinson M F. J Biol Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- 40.Li S L, Leonard D, Wilkinson M F. J Exp Med. 1997;185:985–992. doi: 10.1084/jem.185.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Maquat L E. EMBO J. 1997;16:826–833. doi: 10.1093/emboj/16.4.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagy E, Maquat L E. Trends Biochem Sci. 1998;23:189–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 43.Cheng J, Belgrader P, Zhou X, Maquat L E. Mol Cell Biol. 1994;14:6317–6325. doi: 10.1128/mcb.14.9.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, J., Sun, X., Qian, Y., LaDuca, J. P. & Maquat, L. E., (1998) Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]
- 45.Carter M S, Li S, Wilkinson M F. EMBO J. 1996;15:5965–5975. [PMC free article] [PubMed] [Google Scholar]
- 46.Long R M, Elliot D J, Stutz F, Rosbash M, Singer R H. RNA. 1995;1:1071–1078. [PMC free article] [PubMed] [Google Scholar]