Abstract
Five percent of live-born human offspring will have a genetic disorder. Of these, 20% are because of germ-line de novo mutations. Several genetic diseases, such as neurofibromatosis and Duchenne muscular dystrophy, are associated with a high percentage of de novo germ-line mutations. Until recently, a direct analysis of spontaneous mutation frequencies in mammalian germ cells has been prevented by technical limitations. We have measured spontaneous mutation frequencies in a lacI transgene by using enriched populations of specific spermatogenic cell types. Similar to previously published results, we observed a lower mutation frequency for seminiferous tubule cell preparations, which contain all stages of spermatogenesis, relative to somatic tissues. We made the unexpected observation of a decline in mutation frequency during spermatogenesis, such that the mutation frequencies of type B spermatogonia and all subsequent stages of spermatogenesis are lower than the frequency for primitive type A spermatogonia. In addition, spermatogenic cells from old mice have significantly increased mutation frequencies compared with spermatogenic cells from young or middle-aged mice. Finally, the mutation frequency was observed to increase during spermiogenesis in postreplicative cell types when spermatogenic cells were obtained from old mice.
From a genetic perspective, germ cells are profoundly different from somatic cells because they carry the genetic information that will direct the development of the next generation, not simply the next daughter cell. Thus, safeguarding the integrity of germ-line DNA might provide evolutionary advantages. Indeed, in mice, mutation frequencies obtained from mixed populations of germ cells are lower than for somatic tissues (1). This was demonstrated by using a transgenic system in which the bacteriophage λ genome carrying the lacI repressor gene and the αlacZ gene from the prokaryotic lac operon was introduced into the mouse genome as a transgene. λ DNA was recovered from genomic DNA preparations by packaging and used to infect a strain of Escherichia coli carrying a lacZ (β-galactosidase) gene, but lacking a functional lacI gene. Mutation of the lacI gene renders a blue plaque on agarose containing the chromogenic substrate 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal). In contrast, other studies using a lacI transgenic mouse (2) or a lacZ transgenic mouse (3) did not report a significant difference in mutation frequencies for spermatogenic cells compared with somatic cells.
Although provocative, interpretation of the results demonstrating a lower mutation frequency for male germ cells is complicated by the fact that adult seminiferous tubules contain a mixture of spermatogenic cell types encompassing all stages of spermatogenesis. Spermatogonia serve as the stem cells for spermatogenesis and undergo mitotic divisions that give rise to cells that will either retain their identity as spermatogonia to maintain the stem cell population or enter meiosis to become primary spermatocytes. Once committed to proceed through meiosis, spermatocytes undergo one round of DNA replication and two rounds of cellular division to generate haploid spermatids. After meiosis, round spermatids undergo spermiogenesis to produce spermatozoa, a process involving significant cellular differentiation in the absence of cell division. Somatic Sertoli cells also are present in the seminiferous epithelium and comprise about 3% of the cells in this tissue.
In the present study, we first examined the spontaneous mutation frequency in a mixed population of seminiferous tubule cells and somatic tissues to determine whether the mutation frequency was lower in germ cells. Second, we examined the mutation frequencies in specific spermatogenic cell types from premeiotic, meiotic, and postmeiotic stages to determine when the generally lower mutation frequency observed in seminiferous tubule cells develops during spermatogenesis. Third, we examined mutation frequencies in spermatogenic cells from mice at various ages to determine whether there is any age-related effect on mutation frequency. Our results indicate that the spontaneous mutation frequency is lower in seminiferous tubule cells compared with somatic tissues. In addition, our results show that the mutation frequency declines during spermatogenesis in young mice as cells progress from mitotic spermatogonia to meiotic spermatogonia. The meiotic cell types in young mice retain the lower mutation frequency throughout the remainder of spermatogenesis. In contrast, the mutation frequency is elevated in spermatogenic cells of old mice and also increases during spermatogenesis in old mice. Here, the increase in mutation frequency was observed in postreplicative meiotic cell types.
MATERIALS AND METHODS
Animals.
Male lacI transgenic mice (C3HeB/FeJ × C57BL/6/lacI F1 hybrids) were obtained from Stratagene’s colony (Taconic Labs) or from in-house breeding regimens. The animals were housed in an American Association for the Accreditation of Laboratory Animal Care-accredited animal facility and fed standard mouse lab chow and water ad libitum. Mice were specific-pathogen-free. Mice were humanely euthanized at the appropriate ages. Organs were rapidly removed and used immediately for germ cell separation procedures or frozen (liver and brain) in liquid nitrogen and stored at −80°C until used for genomic DNA isolation.
Spermatogenic Cell Type Separations.
Sertoli cells and specific populations of spermatogenic cells types were prepared by using a standard Sta Put gradient system as described previously (4, 5). Purities of recovered cells were based on morphological criteria and were ≥85%.
Genomic DNA Isolation.
Genomic DNA was isolated by using the RecoverEase DNA isolation kit according to the manufacturer’s recommendations (Stratagene).
Mutagenesis Assay.
Transpack in vitro packaging extract (Stratagene) was used to recover the lacI-containing lambda shuttle vector from genomic DNA according to the supplier’s recommendations. E. coli SCS-8 cells (Stratagene) were mixed with packaged phage, added to top agarose containing X-gal, and plated on NZY agar assay trays. Trays were visually screened for blue mutant plaques. All putative mutant plaques were cored and replated on fresh X-gal/NZY plates to confirm lacI mutations. Mutation frequency was determined by dividing the number of confirmed mutant lacI genes by the total number of plaque-forming units.
To confirm that the lower mutation frequency obtained for seminiferous tubule cells was not a technical artifact of the isolation procedure, mixing experiments were performed. Testis homogenates prepared from lacI transgenic mice were combined 1:4 (vol/vol) with nontransgenic liver homogenates before DNA was isolated. Conversely, liver homogenates prepared from transgenic mice were combined 1:4 with nontransgenic testis homogenates before DNA isolation. Because the mutation frequencies from this experiment (data not shown) were similar to those obtained without mixing, it does not appear that the low mutation frequency observed for seminiferous tubule cells results from a technical artifact.
It is important to note that mutations produced in the mouse rather than the amount of premutational DNA damage or mutations produced in the E. coli host cells via replication errors in the phage DNA have been measured. DNA adducts generated in the mouse also could lead to the production of mutations during replication of phage DNA; however, these plaques would appear mosaic during confirmation analyses because only one strand of the phage DNA would yield a detectable mutation. Therefore, confirmation of mutations prevented inappropriate scoring of mutations produced in E. coli.
Statistical Methods.
Data were examined as described by Piegorsch et al. (6). Because no excess variability was found between packaging reactions and packaging dates, subsequent analyses used pooled data. The numbers of mutants are described by binomial random variables. Mutation frequencies in more than two cell types or ages were compared by using the χ2 test (7). Pairwise comparisons of mutation frequencies were conducted by using the conditional binomial test (8). We analyzed trends with age by using logistic regression, including in the statistical model both a linear trend with age and a nonlinear trend. Test of hypotheses in logistic regression were made with exact tests (logxact 2.0, Cytel Software Corp., Cambridge, MA).
RESULTS
Spontaneous Mutation Frequencies in Somatic Tissues and Young Adult Seminiferous Tubule Cells.
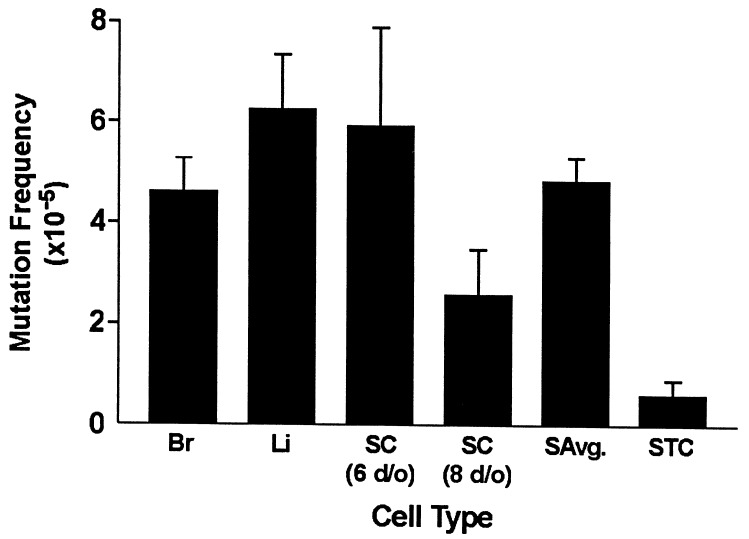
To ensure that our results were comparable with previously published data (9), we first determined mutation frequencies for two somatic tissues, brain and liver (Fig. 1 and Table 1). In addition, the mutation frequency for Sertoli cells, the single somatic cell type within the seminiferous epithelium, also was determined. Fig. 1 shows the average mutation frequencies for the different somatic cell types. These frequencies were similar to those previously reported by Kohler et al. (1) and not significantly different from one another. Thus, an average mutation frequency was calculated for the three somatic tissues and used for subsequent comparisons with germ-cell frequencies. Also consistent with previously published results (1), the mutation frequency of a mixed population of seminiferous tubule cells from young adult mice was determined to be significantly lower than the average somatic frequency (P ≤ 0.0001; Table 1 and Fig. 1).
Figure 1.
Mutation frequencies and standard errors in somatic cells/tissues and enriched seminiferous tubule cell preparations obtained from 60-day-old male mice. The mutation frequencies are significantly lower for seminiferous tubule cells versus somatic cells (P ≤ 0.0001). A test for differences between brain, liver, Sertoli cells (6 days old), and Sertoli cells (8 days old) revealed no significant differences (P = 0.1094). Br, brain; Li, liver; SC, Sertoli cells; SAvg, somatic average; STC, seminiferous tubule cells.
Table 1.
Spontaneous mutation frequencies in murine spermatogenic cell types
| Cell tissue type | No. animals | Total pfu | Mutant plaques | Mutation frequency, × 10−5 ± SE × 10−5 |
|---|---|---|---|---|
| Brain | 5 | 1,043,029 | 48 | 4.6 ± 0.66 |
| Liver | 6 | 527,100 | 33 | 6.3 ± 1.1 |
| Sertoli cell (6 days old) | 9 | 152,300 | 9 | 5.9 ± 2.0 |
| Sertoli cell (8 days old) | 13 | 312,000 | 8 | 2.6 ± 0.91 |
| Somatic cell average | 2,034,429 | 98 | 4.8 ± 0.49 | |
| Seminiferous tubule cells | 4 | 661,920 | 4 | 0.6 ± 0.3 |
| Primitive type A spermatogonia | 6 + 9 | 899,145 | 21 | 2.3 ± 0.5 |
| A + B spermatogonia | 9 | 766,400 | 4 | 0.5 ± 0.3 |
| A spermatogonia | 11 | 570,150 | 11 | 1.9 ± 0.6 |
| B spermatogonia | 11 | 606,200 | 5 | 0.8 ± 0.4 |
| Preleptotene spermatocytes | 5 | 650,450 | 4 | 0.6 ± 0.3 |
| Leptotene + zygotene spermatocytes | 3 | 341,090 | 1 | 0.3 ± 0.3 |
| Young pachytene spermatocytes | 6 | 1,434,780 | 6 | 0.4 ± 0.2 |
| Young round spermatids | 6 | 1,278,300 | 5 | 0.4 ± 0.2 |
| Young spermatozoa | 2 | 637,850 | 5 | 0.8 ± 0.4 |
The total number of plaque-forming units (pfu) analyzed, confirmed mutants, mutation frequencies, and SE are shown for each cell/tissue type.
Spontaneous Mutation Frequencies in Specific Spermatogenic Cell Types.
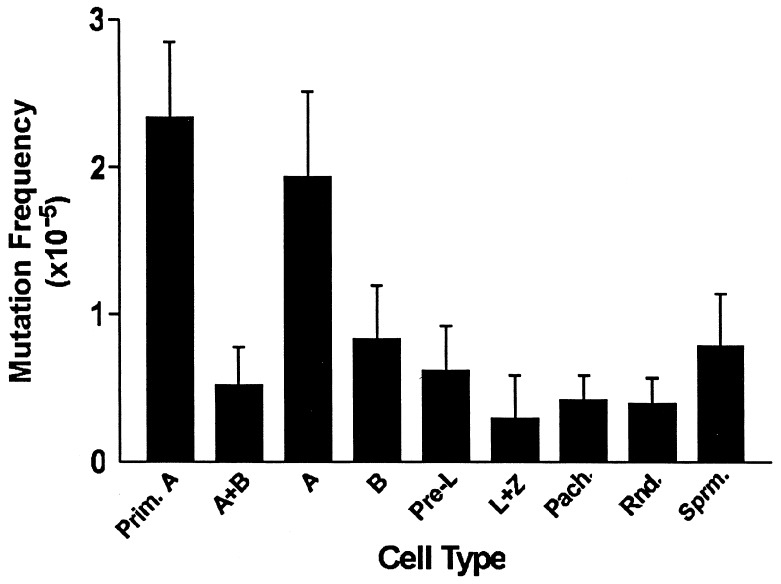
To identify the specific cell types within the seminiferous epithelium that give rise to the low lacI mutation frequency in this tissue, purified populations of various spermatogenic cell types were prepared and the lacI mutation frequencies were determined (Fig. 2 and Table 1). Specifically, we investigated spontaneous mutation frequencies in purified populations of different spermatogenic cell types that occur in the following developmental order in the adult testis: primitive type A spermatogonia, type A spermatogonia, type B spermatogonia, preleptotene primary spermatocytes, leptotene spermatocytes, zygotene spermatocytes, pachytene spermatocytes, round spermatids, and spermatozoa. All of the examined spermatogenic populations exhibited a significantly lower mutation frequency than that observed in somatic cells (P ≤ 0.001; Table 1). The earliest spermatogonial cell type assayed, primitive type A spermatogonia, exhibited the highest mutation frequency and was not significantly different from the frequency of type A spermatogonia (P = 0.3746). The mutation frequency declined significantly in the mitotic derivatives of primitive type A spermatogonia, such that the frequency in a mixed population of types A and B spermatogonia was significantly lower than that observed in primitive type A spermatogonia, with the largest proportion of that decline occurring between type A and type B spermatogonia (Fig. 2 and Table 1).
Figure 2.
Mutation frequencies and standard errors for specific spermatogenic cell types. All cell types tested had a significantly lower mutation frequency than the average somatic mutation frequency (P ≤ 0.001). Furthermore, there was a significant decline in mutation frequency during spermatogenesis between primitive type A spermatogonia and type B spermatogonia (P = 0.02). Prim. A, primitive type A spermatogonia; A, type A spermatogonia; B, type B spermatogonia; Pre-L, preleptotene spermatocytes; L+Z, leptotene plus zygotene spermatocytes; Pach., pachytene spermatocytes; Rnd., round spermatids; Sprm, spermatozoa.
Overall, primitive type A spermatogonia had a greater mutation frequency than mixed seminiferous tubule cells (P = 0.0049), a mixed population of type A + B spermatogonia (P = 0.0017) and type B spermatogonia (P = 0.0200). Each of the later spermatocyte, spermatid, and spermatozoa cell types displayed a relatively low mutation frequency similar to that found in type B spermatogonia (P = 0.6910; Fig. 2).
Spontaneous Mutation Frequencies in Spermatogenic Cell Types Obtained from Old Mice.
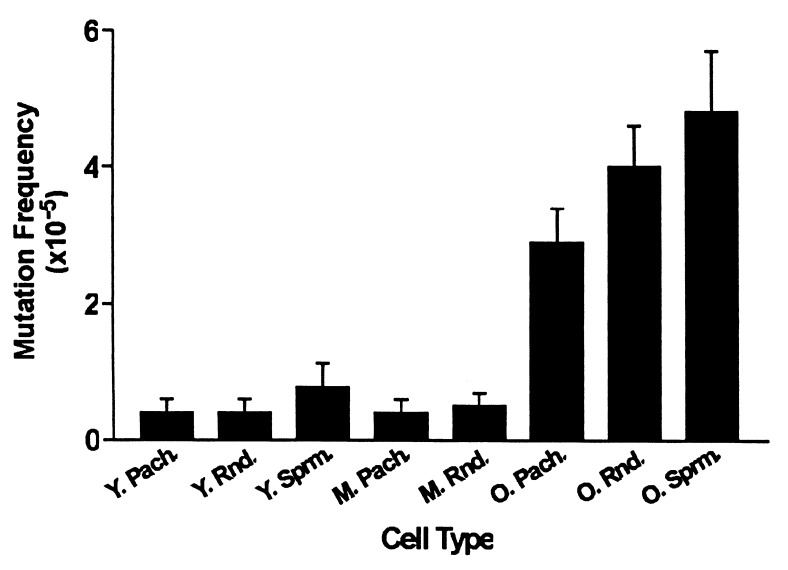
To determine whether the relatively low spontaneous mutation frequencies are maintained in spermatogenic cells as mice age, males carrying the lacI transgene were aged to 15 or 28 months and populations of pachytene spermatocytes, round spermatids, and epididymal/vas deferens spermatozoa were recovered and assayed for mutation frequencies. As shown in Fig. 3 and Table 2, the mutation frequencies were similar in similar spermatogenic cell types from young (ca. 2 months) and middle-aged (ca. 15 months) mice (P ≥ 0.4848); however, they were increased significantly in spermatogenic cells from old (ca. 28 months) mice (P < 0.0001). Thus, the change in mutation frequency with age was nonlinear [nonlinear trend for pachytene spermatocytes (P = 0.0835) and for round spermatids (P = 0.05471)]. A 7-fold increase in mutation frequency was observed between pachytene spermatocytes obtained from old versus young mice, a 10-fold increase for round spermatids, and a 6-fold increase for spermatozoa. These results reveal another phenomenon unique to the old mice: an increase in spontaneous mutation frequency occurs during spermiogenesis and sperm maturation in the aged male reproductive tract. Thus, in old mice, we found that the mutation frequency for any particular spermatogenic cell type was greater than that found in the same cell type in young or middle-aged mice. However, in aged mice we also observed a significant (P = 0.0314) increase in mutation frequency during spermatogenesis, such that the frequency in spermatozoa was greater than that in pachytene spermatocytes.
Figure 3.
Spontaneous mutation frequencies and standard errors are shown for spermatogenic cells obtained from 60-day-old, 15-month-old, and 28-month-old mice. The mutation frequency of old pachytene spermatocytes is greater than that of young spermatocytes (P < 0.0001) and greater than middle-aged pachytene spermatocytes (P < 0.0001). Similarly, the mutation frequency of old round spermatids is greater than young (P < 0.0001) and middle-aged (P < 0.0001) round spermatids. Furthermore, the mutation frequency of old spermatozoa is greater than that of young spermatozoa (P < 0.0001). The mutation frequency was greater in old spermatozoa than in old pachytene spermatocytes (P = 0.0314), but there were no significant differences among young pachytene spermatocytes, round spermatids, and spermatozoa (P = 0.4585). Y. Pach., young pachytene spermatocytes; Y. Rnd., young round spermatids; Y. Sprm., young spermatozoa; M. Pach., middle-aged pachytene spermatocytes; M. Rnd., middle-aged round spermatids; O. Pach., old pachytene spermatocytes; O. Rnd., old round spermatids; O. Sprm., old spermatozoa.
Table 2.
Spontaneous mutation frequencies in murine spermatogenic cell types obtained from mice of various ages
| Cell/tissue type | No. animals | Total pfu | Mutant plaques | Mutation frequency, ×10−5 ± SE × 10−5 |
|---|---|---|---|---|
| Young pachytene spermatocytes | 6 | 1,434,780 | 6 | 0.4 ± 0.2 |
| Young round spermatids | 6 | 1,278,300 | 5 | 0.4 ± 0.2 |
| Young spermatozoa | 2 | 637,850 | 5 | 0.8 ± 0.4 |
| Middle-aged pachytene spermatocytes | 6 | 1,205,290 | 5 | 0.4 ± 0.2 |
| Middle-aged round spermatids | 6 | 1,471,500 | 7 | 0.5 ± 0.2 |
| Old pachytene spermatocytes | 7 | 1,227,410 | 35 | 2.9 ± 0.5 |
| Old round spermatids | 7 | 956,690 | 38 | 4.0 ± 0.6 |
| Old spermatozoa | 5 | 596,255 | 29 | 4.9 ± 0.9 |
The total number of plaque-forming units (pfu) analyzed, confirmed mutants, mutation frequencies, and SE are shown for each sample.
DISCUSSION
Our results support the conclusion that male germ cells have a lower spontaneous mutation frequency than somatic tissues. These frequency data are in accord with mutation frequencies determined indirectly through the specific locus test (10, 11) and frequency data obtained for sperm or testes from lacI and lacZ transgenic mice (1, 12). The only other instance where a decline in mutation frequency has been observed within a lineage was reported by Witkin (13). In that report, the mutation frequency decline (MFD) was demonstrated in prokaryotic organisms after UV irradiation. Although it is relatively easy to understand increases in mutation frequency in a lineage (e.g., accumulation over time), a decrease is more difficult to explain. There is now a partial molecular explanation for the MFD observation described for prokaryotes: the mfd gene encodes a transcription-repair coupling factor (TRCF), which mediates preferential DNA repair of actively transcribed genes after UV exposure (14). However, our data are not explained by coupling transcription with DNA repair because the lacI transgene is not transcribed. Furthermore, our methodology directly measured fixed mutations. Consequently, an increased level of DNA repair could not explain the decline in mutation frequency observed in our studies, because the current understanding of DNA repair pathways does not provide a mechanism for recognition of fixed mutations.
We suggest that multiple mechanisms may contribute to the reduction in mutation frequency we observe in mouse spermatogenic cells. First, germ cells appear to be maintained in a relatively protected state throughout development. This would explain the initially low mutation frequency in primitive type A spermatogonia. Second, an additional quality-control mechanism appears to be operating during the premeiotic stages of spermatogenesis, such that only those cells with particularly low mutation frequencies are allowed to proceed into meiosis. This notion is supported by the observation that significant apoptosis occurs between type A and type B spermatogonia (15–18), a timing that is concordant with the observed decline in mutation frequency. Our results may suggest that spermatogonia bearing relatively higher mutation frequencies are more likely to be included among the apoptotic cells than are those with low mutation frequencies. A mechanism to explain this could involve an active checkpoint whereby spermatogonia are screened for mutations via sentinel genes, specific biomarkers that coincide with elevated mutation frequencies, or it could be that within seminiferous tubules there exists a select subpopulation of germ cells that maintain a reduced mutation frequency throughout development and are preferentially predestined to proceed through spermatogenesis.
It has been reported that the lacI transgene has a high spontaneous incidence of mutagenesis (19). Because the majority of spontaneous lacI mutations have been identified as GC → AT transition mutations that occur predominantly at CpG sequences, it has been hypothesized that these particular mutations may result from the deamination of 5-methylcytosine residues in CpG dinucleotides (1). It will be interesting to determine the nature of the mutations obtained in our study and to determine whether the mutations correlate with methylation changes in the lacI transgene during spermatogenesis, particularly given that many human hereditary diseases appear to result from transition mutations at CpG dinucleotides (20). Thus, the lacI transgenic mouse system may be an extremely useful model for gaining insight into the propagation of transition mutations at CpG dinucleotides.
In humans, 5% of live-born offspring have a genetic disorder (21) and 20% of these are because of de novo mutations in the germ line (22, 23). Some genetic diseases, such as neurofibromatosis (24) and Duchenne muscular dystrophy (25), have been shown to correlate with a high proportion of de novo germ-line mutations. In addition, several autosomal-dominant genetic diseases, such as achondroplasia and hemophilia A, are associated with a high frequency of de novo mutations and are correlated with a higher percentage of paternal mutations (26). Furthermore, de novo mutations in some autosomal-dominant diseases, including achondroplasia, Marfan syndrome, and Apert syndrome (26), correlate with increased paternal age. At least two factors may contribute to these male-specific mutation phenomena. First, spermatogonial stem cells span the majority of the life in males, allowing a greater opportunity for exposure of stem cells to DNA-damaging agents and an accumulation of mutations simply by longevity. Second, male germ cells undergo more cycles of DNA replication and cell division to produce mature gametes than do female germ cells. This provides more opportunities to fix mutations during DNA replication (27).
The nonlinear increase in mutation frequency we detected during murine spermatogenesis with increased age appears to be unique and suggests an age-related breakdown of a mechanism operating solely in germ cells. This is in contrast to the gradual accumulation of mutations that appears to account for the age-related increase in spontaneous mutation frequencies observed for somatic tissues. For example, splenic DNA from lacI transgenic mice was analyzed for mutations at various ages from birth to 24 months old, and a gradual, linear, 4-fold increase in mutation frequency was noted with increasing age (28). In humans, a decrease in DNA-repair capacity of UV products has been demonstrated in peripheral lymphocytes with increased age (29). Again, there was a linear relationship between age and decreased repair capacity. Although repetitive cell divisions may account for some of the increase in mutation frequency observed in spermatogenic cells from old mice, it cannot account for the increase detected between pachytene spermatocytes and spermatozoa because DNA replication does not occur between these stages. Indeed, our results suggest that subsequent to the decline in mutation frequency that occurs in spermatogonia, there are normally additional functioning mechanisms that operate to maintain the low frequency in meiotic and postmeiotic spermatogenic cells, but that these mechanisms lose fidelity in old mice.
Thus, our data lead to the suggestion that, with respect to mutagenesis, the germ line in young males is maintained in a protected state relative to somatic cells. In addition, there appears to be a stage-specific checkpoint at which only those spermatogonia with comparatively low mutation frequencies are allowed to progress into meiotic prophase and through spermatogenesis. Further characterization of the mechanisms involved in the age-related increase in mutation frequency in spermatogenic cells will contribute to an enhanced understanding of the etiology of birth defects arising from paternal de novo germ-line mutations.
Acknowledgments
This research was supported by Grants ES05798 (from the National Institute on Environmental Health Sciences), AG13560 (from the National Institute on Aging), and CA61335 (from the National Cancer Institute) to C.A.W.; RR12253 from the National Center for Research Resources to R.B.W.; and HD23126 from the National Institute of Child Health and Human Development, National Institutes of Health to J.R.M.; and by the Dorothy Coker Research Fellowship and Grant AG000205 from the National Institute on Aging, National Institutes of Health to G.W.I.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Kohler S W, Provost G S, Fieck A, Kretz P L, Bullock W O, Sorge J A, Putman D L, Short J M. Proc Natl Acad Sci USA. 1991;88:7958–7962. doi: 10.1073/pnas.88.18.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishino H, Buettner V L, Haavik J, Schaid D J, Sommer S S. Environ Mol Mutagen. 1996;28:299–312. doi: 10.1002/(SICI)1098-2280(1996)28:4<299::AID-EM2>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 3.Douglas G R, Gingerich J D, Gossen J A, Bartlett S A. Mutagenesis. 1994;9:451–458. doi: 10.1093/mutage/9.5.451. [DOI] [PubMed] [Google Scholar]
- 4.Bellve A R, Cavicchia J C, Millete C F, O’Brien D A, Bhatnagar Y M, Dym M. J Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romrell L J, Bellve A R, Fawcett D W. Dev Biol. 1976;49:119–131. doi: 10.1016/0012-1606(76)90262-1. [DOI] [PubMed] [Google Scholar]
- 6.Piegorsch W W, Lockhart A C, Margolin B H, Tindall K R, Gorelick N J, Short J M, Carr G J, Thompson E D, Shelby M D. Environ Mol Mutagen. 1994;23:17–31. doi: 10.1002/em.2850230105. [DOI] [PubMed] [Google Scholar]
- 7.Snedecor G W, Cochran W G. Statistical Methods. Ames, IA: Iowa State Univ. Press; 1967. pp. 250–253. [Google Scholar]
- 8.Margolin B H, Collings B J, Mason J M. Environ Mutagen. 1983;5:705–716. doi: 10.1002/em.2860050509. [DOI] [PubMed] [Google Scholar]
- 9.Provost G S, Kretz P L, Hamner R T, Matthews C D, Rogers B J, Lundberg K S, Dycaico M J, Short J. Mutat Res. 1993;288:133–149. doi: 10.1016/0027-5107(93)90215-2. [DOI] [PubMed] [Google Scholar]
- 10.Ehling U H, Neuhauser A. Mutat Res. 1979;59:245–256. doi: 10.1016/0027-5107(79)90163-5. [DOI] [PubMed] [Google Scholar]
- 11.Russell W L, Kelly E M, Hunsicker P R, Bangham J W, Maddux S C, Phipps E L. Proc Natl Acad Sci USA. 1979;76:5818–5819. doi: 10.1073/pnas.76.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katoh M, Inomata T, Horiya N, Suzuki F, Shida T, Ishioka K, Shibuya T. Mutat Res. 1994;341:17–28. doi: 10.1016/0165-1218(94)90020-5. [DOI] [PubMed] [Google Scholar]
- 13.Witkin E M. Cold Spring Harbor Symp Quant Biol. 1956;21:123–140. doi: 10.1101/sqb.1956.021.01.011. [DOI] [PubMed] [Google Scholar]
- 14.Drapkin R, Sancar A, Reinberg D. Cell. 1994;77:9–12. doi: 10.1016/0092-8674(94)90228-3. [DOI] [PubMed] [Google Scholar]
- 15.Allan D L, Harmon B V, Kerr J F R. In: Perspectives on Mammalian Cell Death. Potten C S, editor. London: Oxford Univ. Press; 1987. pp. 229–258. [Google Scholar]
- 16.Huckins C, Oakberg E F. Anat Rec. 1978;192:519–528. doi: 10.1002/ar.1091920406. [DOI] [PubMed] [Google Scholar]
- 17.Mori C, Nakamura N, Dix D J, Fujoika M, Nakagawa S, Shiota K, Eddy E M. Dev Dyn. 1997;208:125–136. doi: 10.1002/(SICI)1097-0177(199701)208:1<125::AID-AJA12>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 18.Oakberg E F. Am J Anat. 1956;99:391–409. doi: 10.1002/aja.1000990303. [DOI] [PubMed] [Google Scholar]
- 19.Skopek T R, Kort K L, Marino D R. Environ Mol Mutagen. 1995;26:9–15. doi: 10.1002/em.2850260103. [DOI] [PubMed] [Google Scholar]
- 20.Cooper D N, Krawczak M. Hum Genet. 1990;85:55–74. doi: 10.1007/BF00276326. [DOI] [PubMed] [Google Scholar]
- 21.Hall J G, Powers E K, McIlvaine R T, Ean V H. Am J Med Genet. 1978;1:417–436. doi: 10.1002/ajmg.1320010405. [DOI] [PubMed] [Google Scholar]
- 22.Nelson K, Holmes L B. N Engl J Med. 1989;320:19–23. doi: 10.1056/NEJM198901053200104. [DOI] [PubMed] [Google Scholar]
- 23.Crow J F, Denniston C. Science. 1981;212:888–893. doi: 10.1126/science.7233180. [DOI] [PubMed] [Google Scholar]
- 24.Riccardi V M, Dobson C E, Charkraborty R, Bontke C. Am J Med Genet. 1984;18:169–176. doi: 10.1002/ajmg.1320180121. [DOI] [PubMed] [Google Scholar]
- 25.Harper P S. In: The Metabolic Basis of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 1990. pp. 2869–2902. [Google Scholar]
- 26.Vogel F, Rathenberg R. Adv Hum Genet. 1975;4:223–318. doi: 10.1007/978-1-4615-9068-2_4. [DOI] [PubMed] [Google Scholar]
- 27.Crow J F. Proc Natl Acad Sci USA. 1997;94:8380–8386. doi: 10.1073/pnas.94.16.8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee A T, DeSimone C, Cerami A, Bucala R. FASEB J. 1994;8:545–550. doi: 10.1096/fasebj.8.8.8181674. [DOI] [PubMed] [Google Scholar]
- 29.Wei Q, Matanoski G M, Farmer E R, Hedayati M A, Grossman L. Proc Natl Acad Sci USA. 1993;90:1614–1618. doi: 10.1073/pnas.90.4.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]