Abstract
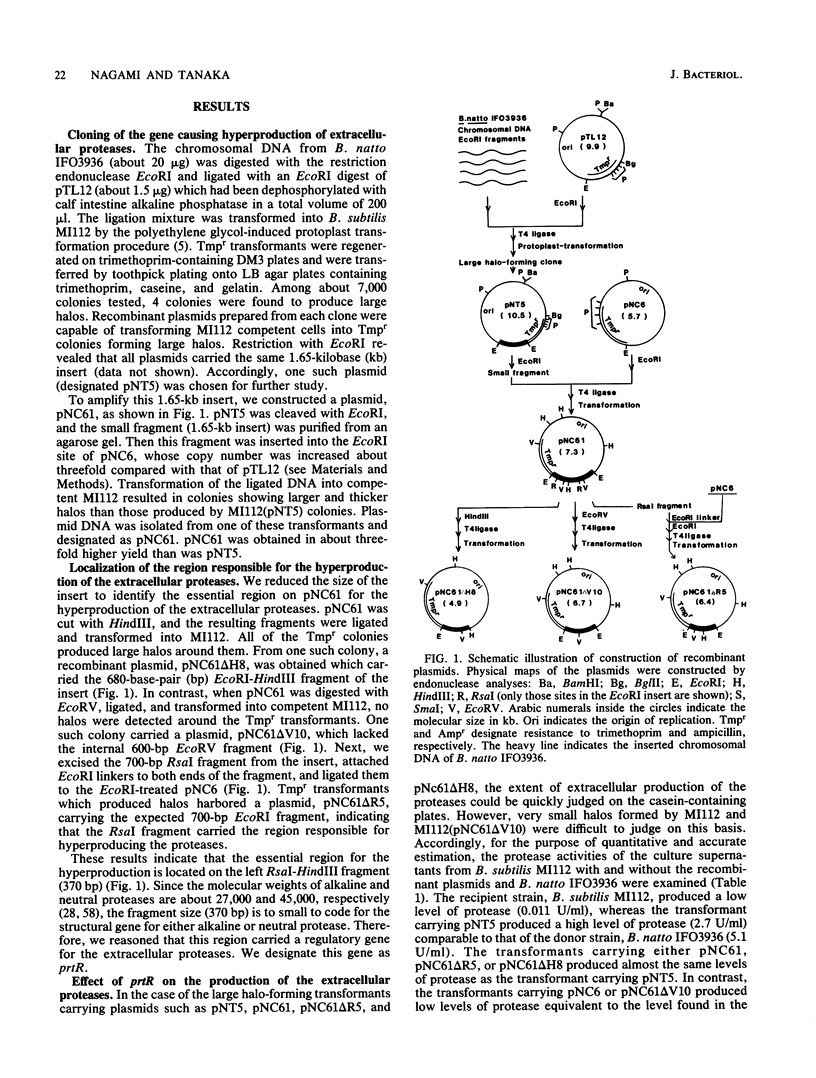
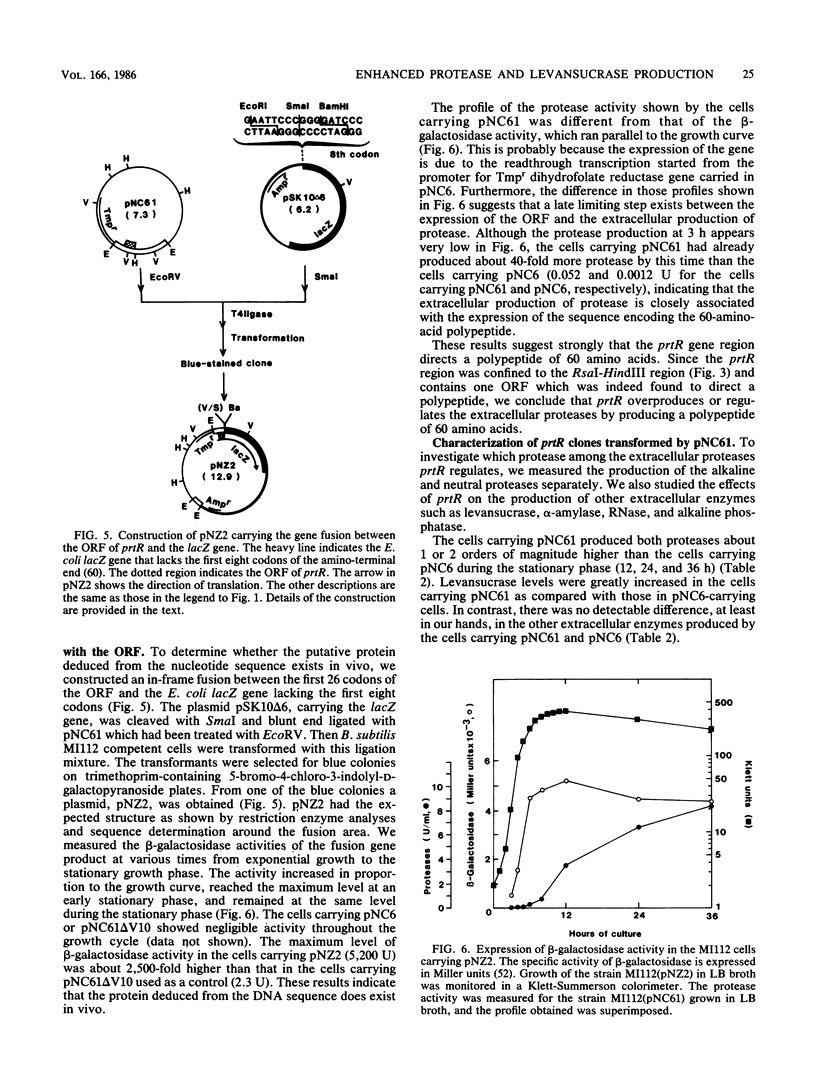
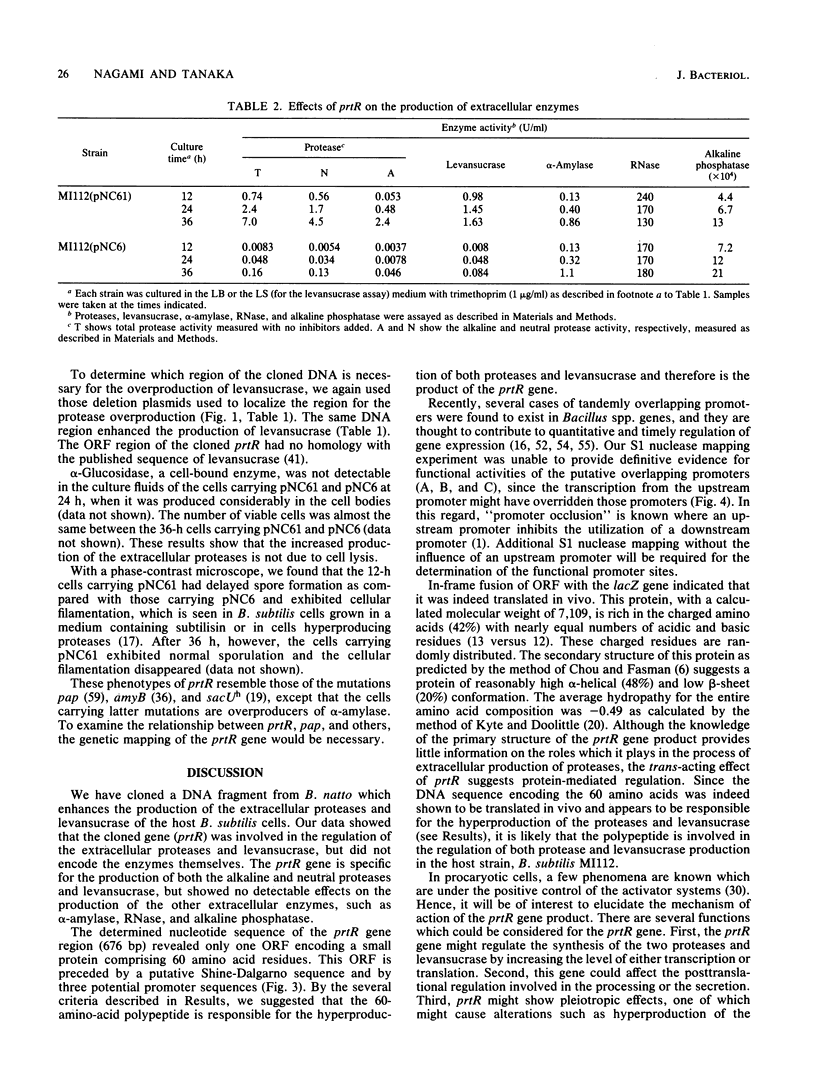
A DNA fragment from Bacillus natto IFO3936 has been cloned which enhances the production of both extracellular alkaline and neutral proteases in Bacillus subtilis. The DNA sequence analysis around the gene responsible for the hyperproduction, prtR, revealed one open reading frame (comprising 60 amino acid residues) which was bounded by potential transcriptional and translational regulatory signals in its preceding and following regions. This open reading frame was not homologous to the published sequences of the structural genes of the two proteases. The calculated molecular weight (7,109) of the polypeptide predicted from the DNA sequence is much smaller than those of the two proteases, indicating that the gene product is distinct from those enzymes. In-frame fusion between the N-terminal region of the coding sequence and the lacZ gene of Escherichia coli demonstrated that the coding region was indeed translated in vivo. By deletion analysis it was suggested that prtR was the structural gene for the 60-amino-acid polypeptide. Cells carrying a prtR plasmid secreted both proteases 40 to 400 times more than the cells carrying the vector alone. Furthermore, it was found that prtR also enhanced the production of levansucrase by 1 or 2 orders of magnitude. There was no difference, however, in the amount of the other extracellular enzymes such as alpha-amylase, RNase, and alkaline phosphatase. These results indicate that prtR is specific for the hyperproduction of the proteases and levansucrase.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Promoter occlusion: transcription through a promoter may inhibit its activity. Cell. 1982 Jul;29(3):939–944. doi: 10.1016/0092-8674(82)90456-1. [DOI] [PubMed] [Google Scholar]
- Aiba S., Kitai K., Imanaka T. Cloning and Expression of Thermostable alpha-Amylase Gene from Bacillus stearothermophilus in Bacillus stearothermophilus and Bacillus subtilis. Appl Environ Microbiol. 1983 Nov;46(5):1059–1065. doi: 10.1128/aem.46.5.1059-1065.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert E., Klier A., Rapoport G. Cloning and expression in Escherichia coli of the regulatory sacU gene from Bacillus subtilis. J Bacteriol. 1985 Mar;161(3):1182–1187. doi: 10.1128/jb.161.3.1182-1187.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Cohen S. N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979 Jan 5;168(1):111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Dancer B. N., Mandelstam J. Production and possible function of serine protease during sporulation of Bacillus subtilis. J Bacteriol. 1975 Feb;121(2):406–410. doi: 10.1128/jb.121.2.406-410.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman M. Z., Chamberlin M. J. Developmental and genetic regulation of Bacillus subtilis genes transcribed by sigma 28-RNA polymerase. Cell. 1983 Nov;35(1):285–293. doi: 10.1016/0092-8674(83)90231-3. [DOI] [PubMed] [Google Scholar]
- Gitt M. A., Wang L. F., Doi R. H. A strong sequence homology exists between the major RNA polymerase sigma factors of Bacillus subtilis and Escherichia coli. J Biol Chem. 1985 Jun 25;260(12):7178–7185. [PubMed] [Google Scholar]
- Higerd T. B., Hoch J. A., Spizizen J. Hyperprotease-producing mutants of Bacillus subtilis. J Bacteriol. 1972 Nov;112(2):1026–1028. doi: 10.1128/jb.112.2.1026-1028.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J. A. Genetics of bacterial sporulation. Adv Genet. 1976;18:69–98. doi: 10.1016/s0065-2660(08)60437-x. [DOI] [PubMed] [Google Scholar]
- Johnson W. C., Moran C. P., Jr, Losick R. Two RNA polymerase sigma factors from Bacillus subtilis discriminate between overlapping promoters for a developmentally regulated gene. Nature. 1983 Apr 28;302(5911):800–804. doi: 10.1038/302800a0. [DOI] [PubMed] [Google Scholar]
- Jolliffe L. K., Doyle R. J., Streips U. N. Extracellular proteases modify cell wall turnover in Bacillus subtilis. J Bacteriol. 1980 Mar;141(3):1199–1208. doi: 10.1128/jb.141.3.1199-1208.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura F., Doi R. H. Construction of a Bacillus subtilis double mutant deficient in extracellular alkaline and neutral proteases. J Bacteriol. 1984 Oct;160(1):442–444. doi: 10.1128/jb.160.1.442-444.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst F., Pascal M., Lepesant-Kejzlarova J., Lepesant J. A., Billault A., Dedonder R. Pleiotropic mutations affecting sporulation conditions and the syntheses of extracellular enzymes in Bacillus subtilis 168. Biochimie. 1974;56(11-12):1481–1489. doi: 10.1016/s0300-9084(75)80270-7. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus beta-lactamase gene. J Biol Chem. 1981 Nov 10;256(21):11283–11291. [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Murphy N., McConnell D. J., Cantwell B. A. The DNA sequence of the gene and genetic control sites for the excreted B. subtilis enzyme beta-glucanase. Nucleic Acids Res. 1984 Jul 11;12(13):5355–5367. doi: 10.1093/nar/12.13.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest F. G. Extracellular enzyme synthesis in the genus Bacillus. Bacteriol Rev. 1977 Sep;41(3):711–753. doi: 10.1128/br.41.3.711-753.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUSHIZKY G. W., GRECO A. E., HARTLEY R. W., Jr, SOBER H. A. STUDIES ON B. SUBTILIS RIBONUCLEASE. I. CHARACTERIZATION OF ENZYMATIC SPECIFICITY. Biochemistry. 1963 Jul-Aug;2:787–793. doi: 10.1021/bi00904a028. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider E., Blundell M., Kennell D. Translation and mRNA decay. Mol Gen Genet. 1978 Apr 6;160(2):121–129. doi: 10.1007/BF00267473. [DOI] [PubMed] [Google Scholar]
- Sekiguchi J., Takada N., Okada H. Genes affecting the productivity of alpha-amylase in Bacillus subtilis Marburg. J Bacteriol. 1975 Feb;121(2):688–694. doi: 10.1128/jb.121.2.688-694.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotsu H., Kawamura F., Kobayashi Y., Saito H. Early sporulation gene spo0F: nucleotide sequence and analysis of gene product. Proc Natl Acad Sci U S A. 1983 Feb;80(3):658–662. doi: 10.1073/pnas.80.3.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M. L., Ferrari E. Replacement of the Bacillus subtilis subtilisin structural gene with an In vitro-derived deletion mutation. J Bacteriol. 1984 May;158(2):411–418. doi: 10.1128/jb.158.2.411-418.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M., Kunst F., Dedonder R. Mapping of mutations affecting synthesis of exocellular enzymes in Bacillus subtilis. Identity of the sacUh, amyB and pap mutations. Mol Gen Genet. 1976 Nov 17;148(3):281–285. doi: 10.1007/BF00332902. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Le Coq D., Aymerich S., Gonzy-Tréboul G., Gay P. The DNA sequence of the gene for the secreted Bacillus subtilis enzyme levansucrase and its genetic control sites. Mol Gen Genet. 1985;200(2):220–228. doi: 10.1007/BF00425427. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Kawano N. Cloning vehicles for the homologous Bacillus subtilis host-vector system. Gene. 1980 Jul;10(2):131–136. doi: 10.1016/0378-1119(80)90130-4. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Koshikawa T. Isolation and characterization of four types of plasmids from Bacillus subtilis (natto). J Bacteriol. 1977 Aug;131(2):699–701. doi: 10.1128/jb.131.2.699-701.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Uehara H., Yamane K., Maruo B. Thermosensitive, extracellular neutral proteases in Bacillus subtilis: isolation, characterization, and genetics. J Bacteriol. 1979 Aug;139(2):583–590. doi: 10.1128/jb.139.2.583-590.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara H., Yoneda Y., Yamane K., Maruo B. Regulation of neutral protease productivity in Bacillus subtilis: transformation of high protease productivity. J Bacteriol. 1974 Jul;119(1):82–91. doi: 10.1128/jb.119.1.82-91.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasantha N., Thompson L. D., Rhodes C., Banner C., Nagle J., Filpula D. Genes for alkaline protease and neutral protease from Bacillus amyloliquefaciens contain a large open reading frame between the regions coding for signal sequence and mature protein. J Bacteriol. 1984 Sep;159(3):811–819. doi: 10.1128/jb.159.3.811-819.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. Z., Doi R. H. Overlapping promoters transcribed by bacillus subtilis sigma 55 and sigma 37 RNA polymerase holoenzymes during growth and stationary phases. J Biol Chem. 1984 Jul 10;259(13):8619–8625. [PubMed] [Google Scholar]
- Wells J. A., Ferrari E., Henner D. J., Estell D. A., Chen E. Y. Cloning, sequencing, and secretion of Bacillus amyloliquefaciens subtilisin in Bacillus subtilis. Nucleic Acids Res. 1983 Nov 25;11(22):7911–7925. doi: 10.1093/nar/11.22.7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H. C., Schnepf H. E., Whiteley H. R. Transcriptional and translational start sites for the Bacillus thuringiensis crystal protein gene. J Biol Chem. 1983 Feb 10;258(3):1960–1967. [PubMed] [Google Scholar]
- Wong S. L., Price C. W., Goldfarb D. S., Doi R. H. The subtilisin E gene of Bacillus subtilis is transcribed from a sigma 37 promoter in vivo. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1184–1188. doi: 10.1073/pnas.81.4.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M. Y., Ferrari E., Henner D. J. Cloning of the neutral protease gene of Bacillus subtilis and the use of the cloned gene to create an in vitro-derived deletion mutation. J Bacteriol. 1984 Oct;160(1):15–21. doi: 10.1128/jb.160.1.15-21.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda Y., Maruo B. Mutation of Bacillus subtilis causing hyperproduction of alpha-amylase and protease, and its synergistic effect. J Bacteriol. 1975 Oct;124(1):48–54. doi: 10.1128/jb.124.1.48-54.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber P., Losick R. Use of a lacZ fusion to study the role of the spoO genes of Bacillus subtilis in developmental regulation. Cell. 1983 Nov;35(1):275–283. doi: 10.1016/0092-8674(83)90230-1. [DOI] [PubMed] [Google Scholar]