Abstract
Previous work has shown that, in the large genomes of three Gramineae [rice, maize, and barley: 415, 2,500, and 5,300 megabases (Mb), respectively] most genes are clustered in long DNA segments (collectively called the “gene space”) that represent a small fraction (12–24%) of nuclear DNA, cover a very narrow (0.8–1.6%) GC range, and are separated by vast expanses of gene-empty sequences. In the present work, we have analyzed the small (ca. 120 Mb) nuclear genome of Arabidopsis thaliana and shown that its organization is drastically different from that of the genomes of Gramineae. Indeed, (i) genes are distributed over about 85% of the main band of DNA in CsCl and cover an 8% GC range; (ii) ORFs are fairly evenly distributed in long (>50 kb) sequences from GenBank that amount to about 10 Mb; and (iii) the GC levels of protein-coding sequences (and of their third codon positions) are correlated with the GC levels of their flanking sequences. The different pattern of gene distribution of Arabidopsis compared with Gramineae appears to be because the genomes of the latter comprise (i) many large gene-empty regions separating gene clusters and (ii) abundant transposons in the intergenic sequences of gene clusters. Both sequences are absent or very scarce in the Arabidopsis genome. These observations provide a comparative view of angiosperm genome organization.
Previous investigations on the nuclear genomes of angiosperms showed that (i) they are characterized by a compositional compartmentalization (1–3) and (ii) that the vast majority of genes from three Gramineae (maize, rice, and barley) are clustered in long DNA stretches that are separated by vast expanses of gene-empty DNA (4, 5). The long gene clusters were collectively called the “gene space” (4, 5). Since the gene space represents only 12–24% of nuclear DNA in the three Gramineae investigated, gene density is four to eight times higher in the gene clusters compared with a uniform gene distribution in those genomes. In the plants tested, the gene space only covers a 0.8–1.6% GC range (GC is the molar fraction of Gua + Cyt in DNA), whereas the DNA bands in CsCl density gradients cover a 20% GC range.
The narrow compositional range of the gene space is due to the narrow compositional spectrum of intergenic sequences in the long gene clusters. In the case of maize, these sequences are largely formed by transposons (6). Since coding sequences from Gramineae cover a broad compositional range, whereas the gene space covers a very narrow range, a correlation is barely detectable between the composition of coding sequences and that of flanking sequences. Very recent work on a 60-kb stretch from the barley genome (7), although not allowing any general conclusion because it only represents 0.001% of the genome, is in agreement with the gene clustering and the GC level previously described (5) for the gene space of barley. It should also be mentioned that some seed storage protein genes, such as zein genes, are located in DNA stretches lower in GC than those hosting the other protein-encoding genes, whereas ribosomal gene clusters are characterized by a much higher GC level.
The gene distribution of a dicot, pea, was basically similar to that found in Gramineae (A.B. and G.B., in preparation). Indeed, most pea genes are located in a gene space covering a 2% GC range and representing less than 20% of the genome (ribosomal genes being again located in very GC-rich fractions). It appears, therefore, that the presence of a gene space is not an exclusive property of Gramineae (whose genome sizes range from 415 megabases (Mb) for rice to 2,500 Mb for maize and to 5,300 Mb for barley; ref. 8), since it is shared at least by pea, a dicot endowed with a large genome, 5,000 Mb.
The possibility that the presence of a gene space is a property associated with the large amounts (50–80%) of repeated sequences in the genomes mentioned above was investigated here by analyzing the gene distribution in the genome of Arabidopsis thaliana. This dicot has an extremely small genome, about 120 Mb, which contains only about 20%–30% of repeated sequences, half of which are highly repetitive, the other half being moderately repetitive (9). Moreover, the Arabidopsis genome contains very few retrotransposons (the Ta and Athila families occur in 15 and 150 copies, respectively; ref. 10) and is endowed with a very low methylation level (less than 6% of all Cyts are methylated; ref. 11).
MATERIALS AND METHODS
Plant and DNA Preparation and Fractionation.
A. thaliana ecotype Columbia was obtained from the Institut National de la Recherche Agronomique (Versailles, France) and the Institut de Biotechnologie des Plantes (Orsay, France). Nuclear DNA was prepared from etiolated seedlings using the method of Jofuku and Goldberg (12) with minor changes. Some contaminating mitochondrial and chloroplast DNAs were, however, present in the DNA preparation. The average molecular weight of the latter was 50 kb, as determined by pulsed-field gel electrophoresis.
DNA Fractionation and Gene Localization.
Fractionation and localization were performed as already described (5). The GC level of the fractions in which genes were localized was calculated from their buoyant densities.
Probes.
Twenty-five expressed sequence tags were obtained from the Arabidopsis Biological Center (Columbus, OH). These were amplified using universal and reverse primers (Pharmacia) and used as radioactive probes.
Compositional Distribution of Coding Sequences.
Complete coding sequences from Arabidopsis were obtained from GenBank release 103 (October 15, 1997) using the ACNUC system (13). Redundancy was eliminated using the CLEANUP program (14) and by visual inspection. The final data set comprised 2,490 sequences. These also included ORFs found in large DNA sequences (from the Arabidopsis genome project) that were annotated as coding sequences in GenBank. In addition, 89 Arabidopsis sequences larger than 50 kb were analyzed. These represented about 10 Mb of DNA, namely, about 8% of the genome.
RESULTS
Compositional Distribution of DNA.
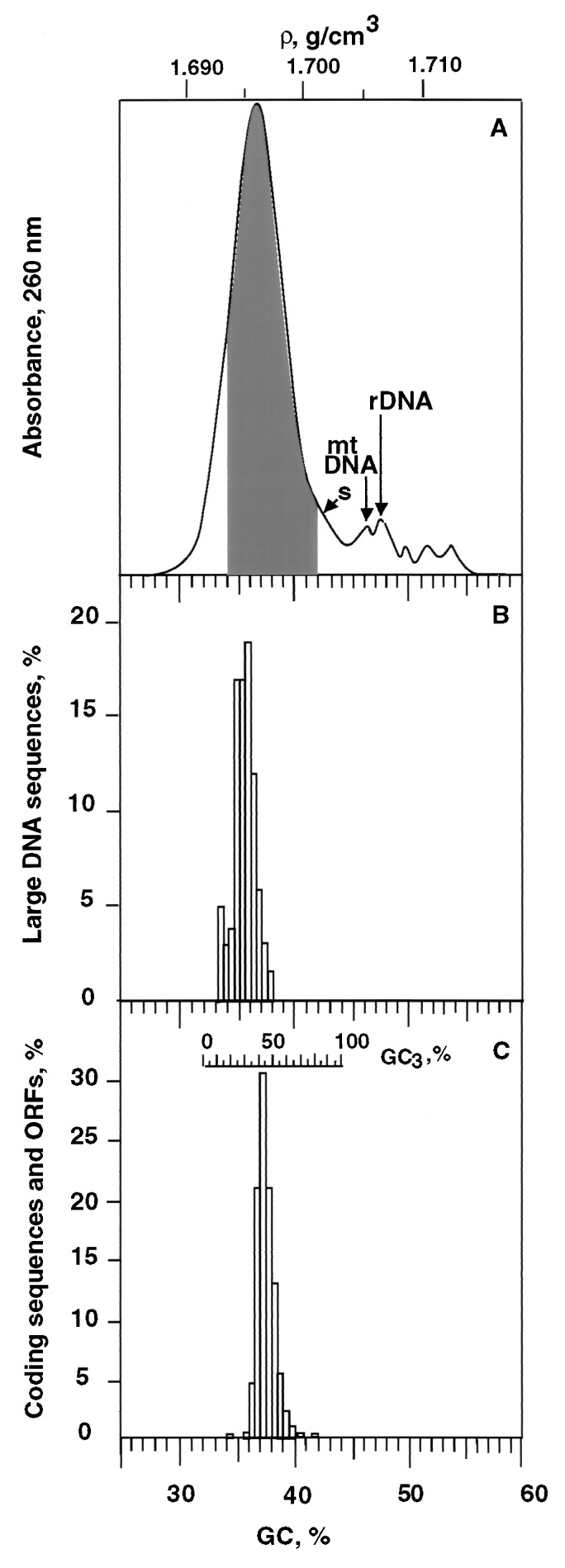
Fig. 1A shows a CsCl profile of Arabidopsis DNA. The whole profile covers a 25 mg/cm3 range of buoyant density corresponding to a range of 25.5% GC. However, the main band only covers a 12 mg/cm3 (15.3% GC) range, from 1.690 to 1.702 g/cm3 (namely, from 30.6 to 42.8% GC) and shows a modal buoyant density of 1.696 g/cm3, corresponding to 36.7% GC.
Figure 1.
(A) Absorbance profile of Arabidopsis nuclear DNA as obtained by centrifugation in a CsCl analytical density gradient. The shoulder (s) may correspond to contaminating chloroplast DNA, the following small peaks to contaminating mitochondrial DNA (ρ = 1.706 g/cm3), rDNA (ρ = 1.707 g/cm3), and to three satellite DNAs (see text). The shaded area corresponds to the DNA fractions containing nuclear protein-encoding genes (see legend of Fig. 2). (B) Compositional distribution of large (>50 kb) GenBank DNA sequences from Arabidopsis. (C) Gene distribution obtained by plotting the relative number of Arabidopsis genes against their GC3 values (top scale); 2,490 sequences from GenBank (release 103; October 15, 1997) were used to construct the histogram. In C, the common GC abscissa of the three plots represents the GC values of the DNA fractions containing the genes (as derived from Fig. 2).
The difference between the GC ranges of the whole profile and main band is due to a high-density shoulder (Fig. 1A, s) and to a series of peaks that comprise contaminating mitochondrial, ribosomal, and satellite DNAs. Indeed, Arabidopsis mitochondrial DNA has a GC level of 44.8% (15) and can, therefore, be identified with the first satellite peak (Fig. 1A). Ribosomal DNA, estimated to represent 6–8% of nuclear DNA (16), has a buoyant density of 1.707 g/cm3, corresponding to 47.9% GC, as indicated by hybridization on DNA fractions (not shown). The shoulder probably corresponds to chloroplast DNA, because chloroplast DNAs from other Brassicaceae show a buoyant density barely higher than nuclear DNA (17). The three other unidentified DNA components are likely to comprise two families of repeated sequences which represent together about 2% of nuclear DNA and are part of the paracentromeric chromatin (16). If all of these heavy DNA components are neglected because they contain no protein-coding nuclear genes, the main CsCl band of Arabidopsis DNA corresponds to about 90% of the nuclear genome, namely, to about 108 Mb.
Compositional Distribution of Large Sequences.
Fig. 1B shows the compositional distribution of 89 sequences larger than 50 kb. This distribution covers a 6% GC range comprised between 33 and 39% GC and is centered on 36.5% GC. This range is narrower than that of the CsCl main band. The mode of the distribution at 36.5% is, however, in excellent agreement with that derived from the CsCl profile, 36.7% GC.
Compositional Distribution of Coding Sequences.
Fig. 1C shows the distribution of the GC3 values (the average GC levels of third codon positions of coding sequences; see the top scale in Fig. 1C) of 2,490 coding sequences (including ORFs; see Materials and Methods). These values range from 18 to 82% and have an average of 45.2%. If coding sequences and ORFs from Arabidopsis contigs are neglected to avoid possible assignment errors in these sequences, the remaining sequences (representing about half of the data set) show an average GC3 of 44.8%, indicating that the possible pollution of coding sequences from contigs does not influence the results of Fig. 1C. In this case, the common GC abscissa in all plots in Fig. 1 represents the GC values of the DNA fractions comprising coding sequences and ORFs (as derived from the correlation of Fig. 2).
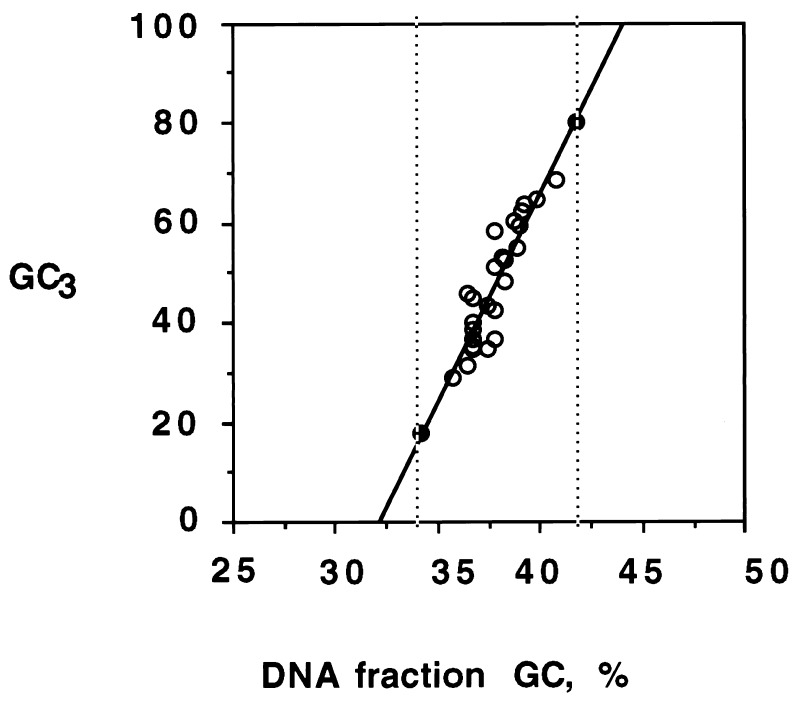
Figure 2.
Plot of GC3 of genes (circles) versus GC values of DNA fractions corresponding to the hybridization peaks (from the data of Table 1). The solid circles represent the two extreme GC3 values of Arabidopsis genes as found in GenBank. The vertical broken lines indicate the GC range of the DNA fractions containing the genes. This was used to define in Fig. 1A (shaded area) the DNA range in which genes are located.
Gene Distribution in the Arabidopsis Genome.
Arabidopsis DNA was fractionated by centrifugation in shallow CsCl gradients (ref. 18; see Fig. 2 of ref. 5 for an example). Twenty-five expressed sequence tags, corresponding to known genes (listed in Table 1), were chosen so as to cover most of the GC3 spectrum of the Arabidopsis genes and ranged from 29 to 69% GC3. These expressed sequence tags were hybridized on the DNA fractions, providing results which were used in constructing Table 1. Table 1 shows that the 25 genes analyzed were localized in DNA fractions having buoyant densities comprised between 1.695 g/cm3 (35.7% GC) and 1.700 g/cm3 (40.8% GC) and covering, therefore, a broad, 5% GC, compositional range.
Table 1.
List of genes localized on CsCl bands of Arabidopsis DNA
| Expressed sequence tag probes* | GC3 | Buoyant density, g/cm3 | GC,† % |
|---|---|---|---|
| ATHB-2 | 28.8 | 1.6950 | 35.71 |
| UBC4 | 31.4 | 1.6957 | 36.42 |
| AIR synthase | 34.6 | 1.6960 | 36.73 |
| KAS-1 | 35 | 1.6967 | 37.44 |
| P5csB | 35.5 | 1.6960 | 36.73 |
| posF21 | 36.8 | 1.6970 | 37.75 |
| Cystathione β-lyase | 37.2 | 1.6967 | 37.44 |
| Amidophosphoribosyltransferase | 38.7 | 1.6960 | 36.73 |
| ACO | 40.1 | 1.6960 | 36.73 |
| CST1-2 | 42.6 | 1.6967 | 37.44 |
| Sat-1 | 43.3 | 1.6967 | 37.44 |
| PAL2 | 44.7 | 1.6955 | 36.22 |
| β-fructofuranosidase | 46 | 1.6975 | 38.26 |
| trpB | 48.4 | 1.6975 | 38.26 |
| TIF4A-1 | 51.1 | 1.6972 | 37.95 |
| S-adenosylmethionine | 52.8 | 1.6975 | 38.26 |
| DWF1 | 53.2 | 1.6975 | 38.26 |
| Oxygen-evolving protein | 55.3 | 1.6981 | 38.87 |
| PUR2 | 58.6 | 1.6977 | 38.46 |
| STP1 | 59.5 | 1.6980 | 38.77 |
| Elongation factor 1α | 60.2 | 1.6977 | 38.46 |
| Geranylphosphate synthase-related protein | 62.1 | 1.6983 | 39.08 |
| ATS3B | 63.7 | 1.6985 | 39.28 |
| Albumin 2S subunit 2 precursor | 64.9 | 1.6986 | 39.38 |
| ROC1 | 68.8 | 1.700 | 40.81 |
Abbreviations of Arabidopsis probes: ATHB-2, DNA-binding protein; UBC4, ubiquitin-conjugating enzyme; AIR synthase, 5′-phosphoribosyl-5-aminoimidazole synthase; KAS-1, 3-ketoacyl carrier protein synthase 1; p5csB, pyrroline-5-carboxylate synthase B; posF21, DNA-binding protein transcription factor; ACO, aconitase; CST1-2, acetolactate synthase; Sat-1, serine acetyltransferase; PAL2, phenylalanine ammonialyase; trpB, tryptophan synthase β subunit; TIF4A-1, eukaryotic translation initiation factor 4A-1; DWF1, Dwarf1; PUR2, glycinamide ribonucleotide synthetase; STP1, glucose transporter; ATS3B, ribulose-1,5-biphosphate carboxylase small subunit; ROC1, cytosolic cyclophilin.
GC of the CsCl fraction corresponding to the hybridization peak.
The results of Table 1 allowed the construction of Fig. 2, which correlates the GC3 values of the genes with the GC values of the DNA fractions in which genes were localized. This correlation has a very good coefficient (r = 0.9) and a high slope (s = 8.2). In fact, coding sequences exhibiting GC3 values as low as 18% and as high as 82% are known in the Arabidopsis genome (see above and Fig. 1C). If these points are introduced in Fig. 2 (solid circles), the GC range of the regions containing genes is extended from 5% (see above) to 8%, namely, from 34% to 42% GC. In turn, if this range is superimposed on the CsCl profile, it defines the DNA range in which the protein-encoding genes are located. This range covers about 85% of the main band, namely, 92 Mb, of Arabidopsis DNA (see Fig. 1A). As expected, the GC values of the same coding sequences are also correlated with the GC values of the corresponding DNA fractions, the slope being, however, only 3.5 (not shown).
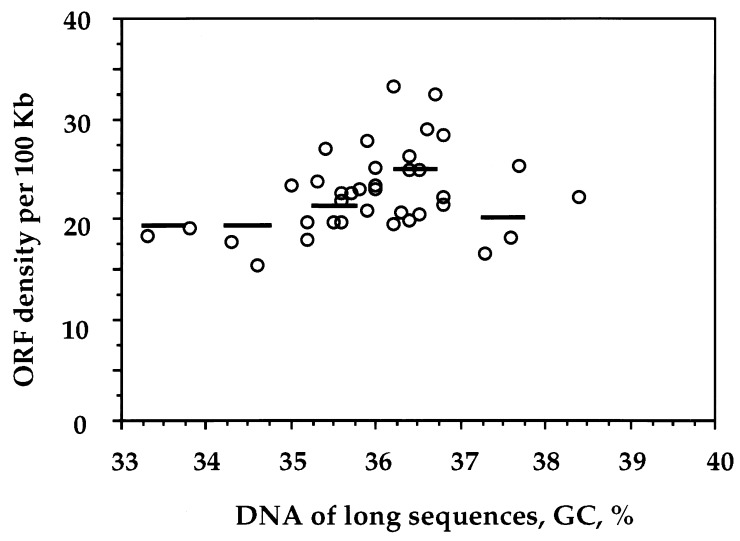
Finally, the distribution of ORFs in the long sequences from Arabidopsis indicated a fairly uniform gene density. Indeed, maximal and minimal values of ORF density were comprised (Fig. 3) within a twofold range (from 15 to 32 ORFs per 100 kb), and the average ORF concentration only varied by ±15% in 1% GC bins.
Figure 3.
ORF density (number of ORFs per 100 kb) in large (>50 kb) DNA segments from Arabidopsis (circles). Average values were also estimated for each 1% GC bin (horizontal bars).
DISCUSSION
Compositional Distribution of DNA Molecules.
As far as the centrifugation in a CsCl density gradient is concerned, Arabidopsis DNA is characterized by a narrower main band (Fig. 1A) compared with the DNAs from Gramineae. Indeed, the GC range of the main band of Arabidopsis DNA is only 12% vs. 20% GC in the case of maize, rice, and barley (5). The modal buoyant density of Arabidopsis DNA, 1.696 g/cm3, corresponds to 36.7% GC, and is significantly lower than the values, 41.4% and 40.3%, that were previously estimated from melting temperature measurements (11) and fluorochrome binding (19), respectively. The 36.7% GC value is in excellent agreement with the modal value, 36.5% GC, found when analyzing long (>50 kb) Arabidopsis sequences present in the GenBank (Fig. 1B). The lower buoyant density of Arabidopsis DNA compared with DNAs from Gramineae (1.7019 g/cm3 for barley, 1.7023 g/cm3 for maize and 1.7026 g/cm3 for rice and wheat; see refs. 1 and 5) account for the fact that rDNA (which has the same buoyant density in all these genomes) appears as a separate peak in Fig. 1A.
The discrepancy in the GC range of the distribution of long (>50 kb) sequences from GenBank (6% GC) and of DNA molecules (12% GC) seems to be essentially due to a lack of representation of GC-poor DNA in the sequences available so far, which only correspond to 8% of the genome. This may be not surprising because the GC-poor DNA is likely to correspond to gene-empty regions of repeated sequences, which have not been cloned or investigated. A spreading of the CsCl band due to the brownian diffusion of the Arabidopsis DNA sample may also contribute, to a small extent, to the discrepancy.
Gene Distribution.
Coding sequences from Arabidopsis (Fig. 1C) are characterized by a wide range of the GC3 distribution (18–82%), in agreement with previous data (1, 4). The average GC level, 45.2%, is higher than the average GC level of main band DNA, about 36.5%, and all points are well above the unity slope line, so that coding sequences stand out, like islands, from the intergenic sequences (increasingly so with increasing GC of coding sequences; this feature was previously found in the human genome; ref. 20).
Arabidopsis genes are distributed over DNA compositional fractions which cover an 8% GC range (Fig. 1A). This GC range is much larger than that found for the gene space in the genomes of Gramineae, 0.8–1.6% GC, or pea, 2% GC, even though these plants have CsCl bands covering a GC range almost twice as broad as the Arabidopsis main band. As expected, Arabidopsis DNA fractions containing genes correspond to a much larger percentage of main band DNA compared with Gramineae (85% vs. 12–24%).
In Arabidopsis there is an evident correlation between the GC3 levels of genes and the GC level of the DNA fractions in which genes are localized (Fig. 2). Because of their very narrow gene spaces, this correlation can barely be detected in Gramineae. The slope of the correlation is much steeper compared with that found (21) in the human genome, 8.2 vs. 2.9. This can be understood if one considers that all Arabidopsis genes are comprised in DNA fragments covering a 8% GC range, whereas human genes are comprised in DNA fragments covering a 25% GC range. The ratio of slopes is very close to the ratio of the ranges, 3.1. Likewise, the slope, 3.5, of GC of coding sequences versus GC of DNA fragments containing them is larger than that, 1.4, exhibited by human coding sequences (20).
As far as gene density is concerned (as derived by ORF density), its variation is very small in Arabidopsis compared with that found in the human genome. Even if extreme values are considered, the overall range of ORF density is only about twofold, a value 10 times lower than that found in the human genome (21). This twofold range of coding sequence densities may be a consequence of the distribution of interspersed repeated sequences, which is known not to be uniform in Arabidopsis (22).
If the number of Arabidopsis genes is assumed to be 20,000, their average density in 85% of the 108-Mb genome comprised in the main band, namely, in 92 Mb, would be about 4.6 kb/gene, a value higher than those, 2–3 kb, found in some small (10–24 kb) regions (23–26) but equal to that reported for a 81-kb region (27) and for a 1.9-Mb contig (28) of the Arabidopsis genome.
Differences Between the Genomes of Gramineae and Arabidopsis.
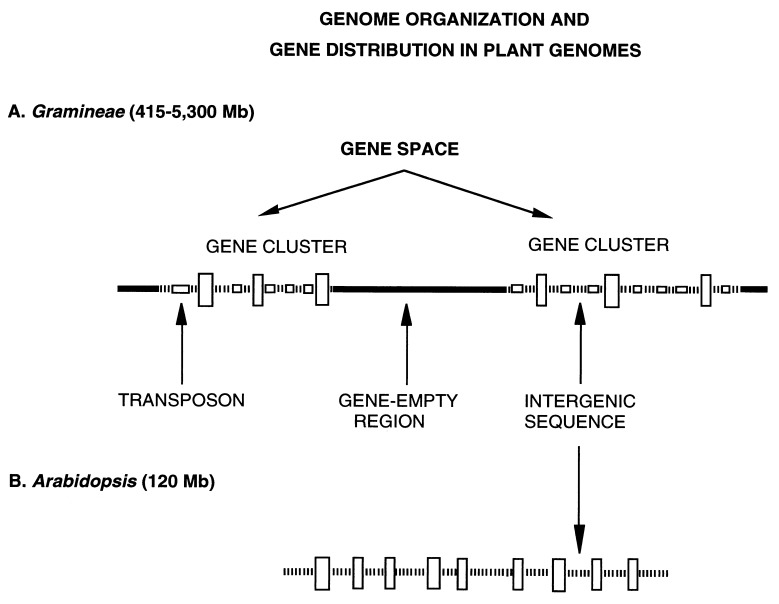
Two major differences distinguish the genomes of Arabidopsis from those of Gramineae. The first one concerns gene distribution. As shown in Fig. 4, in the large genomes, genes are present in long gene clusters separated by long gene-empty regions. The ensemble of long gene clusters forms the gene space which corresponds to 12–24% of the large genomes studied so far. Interestingly, this organization is basically the same over a more than 10-fold range of genome sizes, adding one more basic property to those shared by the genomes of Gramineae, like the collinearity of genetic maps (29) and the compositional correlations among orthologous genes (30). In contrast, in Arabidopsis, genes are fairly evenly distributed over regions amounting to about 85% of the genome, whereas gene-empty regions are greatly reduced and may just consist of the repeated sequences localized in centromeres and telomeres.
Figure 4.
A scheme of genome organization and gene distribution in plant genomes. (A) In the large genomes of Gramineae, genes (large vertical boxes) are present in long gene clusters, which are separated from each other by gene-empty regions formed by repeated sequences (thick solid line). The ensemble of gene clusters forms the gene space. The intergenic sequences are compositionally very homogenous because largely formed by transposons (small horizontal boxes in the intergenic sequences). (B) The small genome of Arabidopsis essentially differs from the genomes of Gramineae because of (i) the disappearance (or very strong reduction) of gene-empty regions; (ii) the practical absence of transposons in intergenic sequences; and (iii) the higher gene density.
The second difference concerns the nature of intergenic sequences. In the large genomes, intergenic sequences within gene clusters (see Fig. 4) are compositionally very homogenous, possibly because largely formed by transposons (6). Indeed, all transposons investigated so far, which include Mu(tator), Ac(tivator) and the majority of Cin4 elements, are exclusively located in the same class of isochores as the ADH-1 gene, i.e., within the gene space (31). This is also true for the many transposons localized near the ADH-1 gene by San Miguel et al. (6). Maize transposable elements appear, therefore, to be located in gene-rich, transcriptionally active regions. Interestingly, this is in agreement with previous observations on transcribed proviral sequences that also integrate into transcriptionally active regions of mammalian genomes (32) and suggested a correlation between integration and transcriptional activity. As a consequence, compositional correlations between coding and flanking sequences are barely detectable. In contrast, in the Arabidopsis genome, which comprises a negligible amount of transposons (10), intergenic sequences are compositionally well correlated with coding sequences.
Two implications of the differences just discussed are (i) that the genomes of Gramineae only differ from the genome of Arabidopsis in having many very large gene-empty regions made up of repeated sequences and large amounts of transposons in the intergenic sequences of their gene clusters and (ii) that the genome of Arabidopsis shares with the human genome the basic property (33) of a compositional correlation between coding sequences and intergenic sequences. These observations provide a comparative view of angiosperm genome organization.
Evolutionary Considerations.
The fact that other members of the Brassicaceae family (which diverged very recently from Arabidopsis; ref. 34) are endowed with large genomes (ref. 19) supports the view that the small genome of Arabidopsis is the result of a marked contraction, in which all (or most) of the large gene-empty regions separating the gene clusters disappeared, as also did most transposons located in intergenic sequences. The opposite viewpoint, that the genome of Arabidopsis was at the origin of the large genomes of other plants, is much less parsimonious. The contraction undergone by the Arabidopsis genome is reminiscent of that shown by the genomes of the fish order Tetraodontiformes. In this case, highly and moderately repeated sequences were greatly reduced (35) compared with the genome of the ancestral order Perciformes, and introns also underwent a contraction (36), especially in GC-poor genes (33). Similar phenomena have been observed in the Arabidopsis introns (N. Carels and G.B., in preparation).
Acknowledgments
We thank the Arabidopsis Biological Resource Center for the gift of probes, Abdellah Hamal (Institut de Biotechnologie des Plantes, Bâtiment 630, Université Paris Sud, F-91405 Orsay, France) for the gift of DNA, Oliver Clay and Gabriel Macaya for discussions, Michel Delseny for communicating results prior to publication, and Nicolas Carels for his help in analytical ultracentrifugation.
ABBREVIATION
- Mb
megabase
References
- 1.Salinas J, Matassi G, Montero L M, Bernardi G. Nucleic Acids Res. 1988;19:5561–5567. [Google Scholar]
- 2.Matassi G, Montero L M, Salinas J, Bernardi G. Nucleic Acids Res. 1989;17:5273–5290. doi: 10.1093/nar/17.13.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Montero L M, Matassi G, Bernardi G. Nucleic Acids Res. 1990;18:1859–1867. doi: 10.1093/nar/18.7.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carels N, Barakat A, Bernardi G. Proc Natl Acad Sci USA. 1995;92:11057–11060. doi: 10.1073/pnas.92.24.11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barakat A, Carels N, Bernardi G. Proc Natl Acad Sci USA. 1997;94:6857–6861. doi: 10.1073/pnas.94.13.6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.San Miguel P, Tikhonov A, Jin Y K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer P S, Edwards K J, Lee M, Avramova Z, Bennetzen J L. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 7.Panstruga R, Büschges R, Piffanelli P, Schulze-Lefert P. Nucleic Acids Res. 1998;26:1056–1062. doi: 10.1093/nar/26.4.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shields R. Nature (London) 1993;365:297–298. [Google Scholar]
- 9.Meyerowitz E M. In: Methods in Arabidopsis Research. Koncz C, Chua N H, Shell J, editors. Singapore: World Scientific; 1992. pp. 11–118. [Google Scholar]
- 10.Thompson H L, Schmit R, Dean C. Nucleic Acids Res. 1996;24:3017–3022. doi: 10.1093/nar/24.15.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leutwiler L S, Hough-Evans B R, Meyerowitz E M. Mol Gen Genet. 1984;194:15–23. [Google Scholar]
- 12.Jofuku K D, Goldberg R B. In: Plant Molecular Biology: A Practical Approach. Shaw C H, editor. Oxford: IRL; 1988. pp. 37–66. [Google Scholar]
- 13.Gouy M, Gautier C, Attimonelli M, Lanave C, Dipaolo G. CABIOS. 1985;1:167–172. doi: 10.1093/bioinformatics/1.3.167. [DOI] [PubMed] [Google Scholar]
- 14.Grillo G, Attimonelli M, Liuni S, Pesole G. Comp App Biosci. 1996;12:1–8. doi: 10.1093/bioinformatics/12.1.1. [DOI] [PubMed] [Google Scholar]
- 15.Unfeld M, Marienfeld J R, Brandt P, Brennicke A. Nat Genet. 1997;15:57–61. doi: 10.1038/ng0197-57. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt R, Putterill J, West J, Cnops G, Robson F, Coupland G, Dean C. Plant J. 1994;5:735–744. doi: 10.1111/j.1365-313x.1994.00735.x. [DOI] [PubMed] [Google Scholar]
- 17.Kirk J T O. In: Handbook of Biochemistry and Molecular Biology. Fasman G D, editor. Boca Raton, FL: CRC Press; 1976. pp. 356–358. [Google Scholar]
- 18.De Sario A, Geigl E-M, Bernardi G. Nucleic Acids Res. 1995;23:4013–4014. doi: 10.1093/nar/23.19.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marie D, Brown S C. Biol Cell. 1993;78:41–51. doi: 10.1016/0248-4900(93)90113-s. [DOI] [PubMed] [Google Scholar]
- 20.Clay O, Caccio S, Zoubak S, Mouchiroud D, Bernardi G. Mol Phylogen Evol. 1996;5:2–12. doi: 10.1006/mpev.1996.0002. [DOI] [PubMed] [Google Scholar]
- 21.Zoubak S, Clay O, Bernardi G. Gene. 1996;174:95–102. doi: 10.1016/0378-1119(96)00393-9. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt R, West J, Love K, Lenehan Z, Lister C, Thompson H, Bouchez D, Dean C. Science. 1995;270:480–483. doi: 10.1126/science.270.5235.480. [DOI] [PubMed] [Google Scholar]
- 23.Le Guen L, Thomas M, Kreis M. Mol Gen Genet. 1994;245:390–396. doi: 10.1007/BF00290120. [DOI] [PubMed] [Google Scholar]
- 24.Aubourg S, Takvorian A, Chéron A, Kreis M, Lecharny A. Gene. 1997;199:241–253. doi: 10.1016/s0378-1119(97)00374-0. [DOI] [PubMed] [Google Scholar]
- 25.Terryn N, Neyt P, De Clercq R, De Keyser A, Van Den Daele H, Ardiles W, Déhais P, Rouzé P, Gielen J, Villarroel R, Montagu M V. FEBS Lett. 1997;416:156–160. doi: 10.1016/s0014-5793(97)01190-3. [DOI] [PubMed] [Google Scholar]
- 26.Van Drunen C M, Oosterling R W, Keultjes G M, Weisbeek P J, Driel R V, Smeekens S C M. Nucleic Acids Res. 1997;25:3904–3911. doi: 10.1093/nar/25.19.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quigley F, Dao P, Cottet A, Mache R. Nucleic Acids Res. 1996;24:4313–4318. doi: 10.1093/nar/24.21.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bevan M, et al. Nature (London) 1998;391:485–488. doi: 10.1038/35140. [DOI] [PubMed] [Google Scholar]
- 29.Bennetzen J L, Freeling M. Trends Genet. 1993;9:259–261. doi: 10.1016/0168-9525(93)90001-x. [DOI] [PubMed] [Google Scholar]
- 30.Carels A, Hatey P, Jabbari K, Bernardi G. J Mol Evol. 1998;45:45–53. doi: 10.1007/pl00006282. [DOI] [PubMed] [Google Scholar]
- 31.Capel J, Montero L M, Marinez-Zapater J M, Salinas J. Nucleic Acids Res. 1993;21:2369–2373. doi: 10.1093/nar/21.10.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zoubak S, Richardson J H, Rynditch A, Höllsberg P, Hafler D A, Boeri E, Lever A M L, Bernardi G. Gene. 1994;143:155–163. doi: 10.1016/0378-1119(94)90091-4. [DOI] [PubMed] [Google Scholar]
- 33.Bernardi G. Annu Rev Genet. 1995;29:445–476. doi: 10.1146/annurev.ge.29.120195.002305. [DOI] [PubMed] [Google Scholar]
- 34.Crepet W L, Feldman G D. Am J Bot. 1991;78:1010–1014. [Google Scholar]
- 35.Pizon V, Cuny G, Bernardi G. Eur J Biochem. 1984;140:25–30. doi: 10.1111/j.1432-1033.1984.tb08062.x. [DOI] [PubMed] [Google Scholar]
- 36.Brenner S, Elgar G, Sandford R, Macrae A, Ventesh B, Aparicio S. Nature (London) 1993;366:265–268. doi: 10.1038/366265a0. [DOI] [PubMed] [Google Scholar]