Abstract
The T lineage repertoire is shaped by T cell receptor (TCR)-dependent positive and negative thymic selection processes. Using TCR-transgenic (N15tg) β2-microglobulin-deficient (β2m−/−) RAG-2−/− H-2b mice specific for the VSV8 (RGYVYQGL) octapeptide bound to Kb, we identified a single weak agonist peptide variant V4L (L4) inducing phenotypic and functional T cell maturation. The cognate VSV8 peptide, in contrast, triggers negative selection. The crystal structure of L4/Kb was determined and refined to 2.1 Å for comparison with the VSV8/Kb structure at similar resolution. Aside from changes on the p4 side chain of L4 and the resulting alteration of the exposed Kb Lys-66 side chain, these two structures are essentially identical. Hence, a given TCR recognizes subtle distinctions between highly related ligands, resulting in dramatically different selection outcomes. Based on these finding and the recent structural elucidation of the N15-VSV8/Kb complex, moreover, it appears that the germ-line Vα repertoire contributes in a significant way to positive selection.
T lymphocytes recognize cell-associated pathogens and tumor antigens bound to class I and class II major histocompatibility complex (MHC) molecules through their clonally distributed αβ T cell receptors (TCRs; refs. 1–3). The understanding of the structure and function of TCRs is thus of theoretical as well as practical importance. Although functional studies long ago documented the dual requirement of specific peptide and MHC molecules in the T cell recognition process (4), structures of TCRs and peptides complexed to MHC molecules (pMHC) only recently have been forthcoming (5–12). As anticipated, the three-dimensional structure of the TCR resembles an antibody Fab fragment such that each of the α and β chains consists of Ig-like variable and constant domains with the hypervariable loops from the two variable domains forming a combining site. This site interacts with the exposed residues of a peptide that sits in the peptide-binding groove of the MHC molecule. The sides of this antigen-presenting platform are composed of two α-helical segments whereas the base consists exclusively of β strands (8–14). Both MHC α-helices interact with the TCR and, indeed, offer a greater recognition surface than the exposed peptide residues (5, 6, 15, 16).
We have focused structural efforts on a class I MHC-restricted αβ TCR derived from a representative cytotoxic T lymphocyte clone, termed N15, with specificity for a vesicular stomatitis virus nuclear octapeptide N52–59 (VSV8) in the context of the Kb mouse class I MHC molecule (17). Prior crystallographic studies of the VSV8/Kb ligand showed that only peptide side chains at positions p1, p4, and p6 were solvent-exposed and, hence, accessible to the TCR (13, 14, 18). Concomitant mutational studies confirmed the importance of these residues in N15 TCR recognition (17, 19). Moreover, comparison of N15 TCR-VSV8/Kb, murine 2C TCR-dEV8/Kb, and human A6 TCR-Tax/HLA-A2 complexes identifies a common docking mode regardless of TCR specificity or species origin in which the TCR variable Vα domain overlays the MHC α2 helix and the Vβ domain overlays the MHC α1 helix (5, 6, 15). As a consequence of this orientation, the CDR1 and CDR3 loops of the TCRα and -β V domains make the major contacts with the peptide whereas the CDR2 loops interact primarily with the MHC α helices.
To investigate in general terms how TCR recognition functions in vivo and, in particular, how the N15 TCR is involved in the process of thymic positive and negative selection, we have established N15 TCR transgenic mice (N15tg) RAG-2−/− H-2b mice (20). On this genetic background, TCR expression is limited to the N15 αβ heterodimer, thereby offering a simplified means for analysis of the N15 TCR throughout T lineage development. Using these mice, we showed previously that intravenous administration of the antigenic VSV8 peptide induced activation of mature N15 T cells in the peripheral lymphoid compartment but mediated negative selection of double-positive (DP) CD4+CD8+ immature thymocytes. The negative selection of the DP thymocytes could be detected within 4 h after VSV8 administration by TUNEL assay or by the presence of activated caspases in N15 fetal thymic organ cultures (FTOC) or adult thymuses (20–22). This process is peptide-specific and independent of peripheral T cell activation (23). In this study, we report on a single altered peptide ligand (APL) variant V4L (L4) of VSV8 that induces the maturation of N15 T cells via positive selection. We then determined a high-resolution crystal structure of the L4 peptide in complex with Kb and identified focal structural differences between the VSV8/Kb and L4/Kb ligands, which induce negative and positive selection of N15 TCR-bearing thymocytes, respectively.
MATERIALS AND METHODS
Animals.
N15 TCR tg RAG-2−/− H-2b mice were constructed as described previously (20). To create the N15+/+RAG-2−/−β2m−/− mice, the N15+/+RAG-2−/− mice were bred with β2m−/− mice that had been backcrossed for 10 generations into C57BL/6 (H-2b) (The Jackson Laboratory). The heterozygous mice from the first generation were intercrossed, and offspring that were RAG-2−/−β2m−/−N15+/+ were utilized as described (22). The expression of the N15 TCR as well as the lack of RAG-2 or β2m gene expression in the knockouts were detected by fluorescence-activated cell sorter (FACS) analysis on peripheral blood cells as explained below. The homozygosity of the N15tg was proven by subsequent breeding analysis. All lines were maintained and bred under sterile barrier conditions at the animal facility of Dana–Farber Cancer Institute.
Flow Cytometric Analysis.
The following mAbs were used: anti-CD4 (H129.19) R phycoerythrin (PE)-labeled and anti-CD8 (53–6.7) Red613-labeled (Life Technologies, Grand Island, NY); anti-N15β [R53 (24)], anti-mouse CD45R/B220 (RA3–6B2) PE-labeled (PharMingen); 5F1.1.24 (anti-mouse H-2Kb) fluorescein isothiocyanate (FITC)-labeled and goat anti-rat IgG FITC-labeled (Caltag, South San Francisco, CA). Thymocytes were triple-stained as follows: cells were incubated with R53 for 30 min followed by FITC-conjugated goat anti-rat antibodies. After washing with PBS containing 2% fetal calf serum (FCS) the cells were restained with PE-anti-CD4 and Red613-anti CD8 mAbs for an additional 30 min in PBS containing 2% FCS and 2% normal rat serum. For RAG-2 and β2m phenotyping, peripheral blood nucleated cells were double stained with PE-anti-mouse CD45R/B220 and FITC-5F1 for 30 min, and, before analysis, red blood cells were eliminated from peripheral blood cells by ammonium chloride lysis. Flow cytometric analysis was performed on a FACScan (Becton Dickinson). Samples were gated on live cells based on forward and side scatter parameters. Data (10,000 events per sample) were collected in list mode by using facscan research software and analyzed by using lysis ii software (Becton Dickinson).
Crystallization, Data Collection, and Structure Determination.
Crystals were grown by hanging-drop vapor-diffusion method. A 10 mg/ml L4/Kb protein solution was mixed up with crystallization buffer of 1.6 M phosphate/2.7% 2-methyl-2,4-pentanediol/0.1 M Hepes, pH 7.0 at equal volume ratio and then equilibrated against the same crystallization buffer. Crystals were grown in two different morphologies, a chunky one and a plate-like one. The former diffracted to about 2 Å, whereas the latter had a larger cell volume and diffracted to 3 Å. The chunky form was used for this high-resolution structure analysis. The crystals were first transferred from the crystallization buffer to cryoprotectant solution, which contains 30% ethylene glycol in the crystallization buffer, and then dipped into liquid nitrogen for rapid freezing. One single frozen crystal of 80 × 70 × 40 μm was used for data collection at Brookhaven NSLS beamline X8C with MAR research imaging plate system at the 1.009-Å wavelength. The crystal belongs to space group P21212 with cell dimension a = 136.7 Å, b = 88.0 Å, and c = 45.2 Å. There is one molecule per asymmetric unit. The data set was reduced and scaled by using programs denzo and scalepack (25). The overall Rsym is 7.6% in 15- to 2.1-Å resolution (see Table 2). The structure was determined by using the molecular replacement method with program amore (26). The search model was taken from the Protein Data Bank, using the same class I Kb molecule loaded with VSV8 (access code 2VAA). The search was done using 15- to 4-Å data only. The solution was extremely clear, with a correlation coefficient value being 66.5 (the next highest value was 38.6). x-plor (27) was used for refinement. Ten percent of reflections was randomly selected as a test set in the refinement. A rigid body refinement followed by alternative cycles of positional and B-factor refinement were carried out. The refinement was started at 15–3 Å and immediately extended to 2.5 Å and, finally, 2.1 Å. Bulk solvent correction was applied. The 2Fo − Fc and Fo − Fc difference maps were calculated with the peptide omitted in Fc calculation. The electron density clearly showed the replacement of Val at p4 position by Leu. A leucine residue was then built in this position of the peptide, and the Kb model was also rebuilt to better fit the density. The model was subjected to further alternative cycles of positional and B-factor refinement. The current Rfree and Rwork are 29.6% and 24.4%, respectively. The final statistics also are given in Table 2.
Table 2.
Crystallographic data
| Data | Value |
|---|---|
| Space group | P21212 |
| Unit cell | a = 136.7 Å, b = 88.0 Å, c = 45.3 Å |
| Resolution range, Å | 15–2.1 |
| Rsym, % | 7.6 (29.0) |
| Completeness, % | 98.8 (98.8) |
| Unique reflections | 32,332 |
| Redundancy | 7.1 (4.0) |
| 〈I〉/〈σ〉 | 15.0 (7.8) |
| Rwork, % | 24.4 (28.1) |
| Rfree, % | 29.6 (29.7) |
| rms bond lengths, Å | 0.006 |
| rms bond angles, ° | 1.398 |
Numbers in parentheses correspond to the last resolution bin, 2.1–2.2 Å.
Peptide Characterization.
Peptide synthesis and analysis were performed by using methods described previously (19). For agonists, the ability of splenocytes derived from N15tg/RAG-2−/− mice to respond to peptide preloaded on irradiated EL-4 cells was tested exactly as described (20). Antagonist peptides for the N15 TCR were identified by using a prepulse assay for inhibition of cytotoxic T lymphocyte lysis. EL-4 cells were labeled with sodium [51Cr]chromate for 1 h at 37°C. A suboptimal concentration of VSV8 (10 pM) was introduced for the duration of the labeling. The cells were washed four times, and 5,000 cells were transferred to each well of a 96-well plate. Test peptides were added at various concentrations. After 1 h at 37°C, 5 × 104 N15 cytotoxic T lymphocytes were added per well. After 4 h, the plates were centrifuged and 100 μl of the supernatant was removed and counted. For positive selection, fetuses of N15tg/RAG-2−/−/β2m−/− mice were dissected at day 15.5 and fetal thymic lobes were cultured with or without 10 μM of the indicated peptide as described (21). Human β2m (Calbiochem) was added to the AIM-V medium (GIBCO/BRL) at 5 μg/ml, and the medium was replaced every 48 h. After 7 days, thymocytes were released from the lobes by pressing through a steel mesh, counted, triple-stained with anti-CD4, anti-CD8, and mAb R53 (anti-N15β chain clonotype), and examined on FACScan. Positive selection was observed by the increase in the percentage of CD8+ single-positive (SP) TCRhigh thymocytes. Negative selection was observed exactly as described (20).
RESULTS AND DISCUSSION
VSV8 Octamer Variants.
TCR-based activation studies using mature T lymphocytes indicate that TCR recognition is exquisitely specific (1–3). Single amino acid changes of even a conservative nature within the antigenic peptide can result in loss of functional activity. Other APLs can function as antagonists or agonists in a clone-specific manner (28–31). The basis of these functional differences among peptides that bind to a given MHC molecule with the same affinity is currently a matter of substantial speculation (32). To further investigate how APLs influence mature T cell activation and thymic selection of N15 TCR-bearing cells, we created a panel of APLs by synthesizing individual VSV8 octamer variants with single amino acid substitutions at the known TCR contact residues. RMA-S stabilization assay demonstrated that all of the APLs bound to Kb with similar affinity (data not shown).
Table 1 shows the sequence variation of individual peptides and summarizes their functional effect on mature peripheral N15+ T cells derived from the N15tg animals. As indicated, none of the variant peptides with substitutions at the p1 or p6 position functioned as agonists to activate mature T lymphocyte proliferation against VSV8-pulsed APCs or lyse VSV8-treated target cells. In addition, all three p1 substitutions (E1, A1, and K1) and the N6-substituted p6 peptide resulted in APLs with antagonist activity in cytotoxicity assay. The ability of conservative substitutions to alter function of the T cells after TCR recognition (for example, the K1 mutation) is striking and, furthermore, consistent with the findings of others studying APLs in both class I and class II TCR-restricted systems (28–31). Substitutions at the VSV8 p4 position yielded several APLs with very different behavior from the p1- and p6-substituted peptides. For example, the I4 peptide functioned as a full agonist, equivalent to the antigenic VSV8 peptide, promoting activation of mature T lymphocytes. On the other hand, the A4 peptide failed to induce any mature T cell response whereas L4 was a weak but clear-cut agonist, requiring a 1,000–10,000 greater concentration in molar terms than VSV8 or I4 to mediate equivalent proliferation or cytotoxic effector function. Note that L4, in contrast to any of the p1 APLs or N6, failed to show antagonist activity.
Table 1.
Effects of Kb binding peptides on mature T cell function and thymic selection in N15tg mice
| Peptide name | Sequence | Agonist | Antagonist | Positive selection | Negative selection |
|---|---|---|---|---|---|
| p1 p4 p6 | |||||
| | | | | |||||
| VSV8 | RGYVYQGL | ■ ■ ■ | □ | □ | ■ ■ ■ |
| E1 | EGYVYQGL | □ | ■ ■ | □ | □ |
| A1 | AGYVYQGL | □ | ■ ■ ■ | □ | □ |
| K1 | KGYVYQGL | □ | ■ ■ ■ | □ | □ |
| L4 | RGYLYQGL | ■ | □ | ■ ■ ■ | ■ |
| I4 | RGYIYQGL | ■ ■ ■ | □ | □ | ■ ■ ■ |
| A4 | RGYAYQGL | □ | □ | □ | □ |
| N6 | RGYVYNGL | □ | ■ ■ ■ | □ | □ |
| E6 | RGYVYNGL | □ | □ | □ | □ |
| A6 | RGYVYAGL | □ | □ | □ | □ |
| OVAp | SIINFEKL | □ | □ | □ | □ |
| SEV9 | FAPGNYPAL | □ | □ | □ | □ |
Agonist, antagonist, positive-selection, and negative-selection activities are as described in Materials and Methods. For agonists, the single, open squares represent no activation of N15 T cell proliferation as judged by stimulation of [3H]thymidine incorporation or N15 CTL effector function at peptide concentrations of 10−4–10−13 M. The single, solid square represents activity in N15 T cell proliferation assays requiring ≥10 μM peptide and, in CTL assay, ≥1 nM. The triple, solid squares represent activity in N15 T cell proliferation assays requiring ≥1 nM and ≥100 pM in CTL assay. For antagonists, the single, open square represents no inhibition of N15 CTL effector function. The double and triple, solid squares represent 50% inhibition of the N15 CTL cytotoxicity at 500 nM peptide and 50 nM, respectively. For positive selection, the single, open square represents no increase of CD8+SP TCRhigh cells in N15tg/RAG-2−/−/β2M−/− FTOC incubated with peptides at concentrations 10−4–10−12 M. The triple, solid square represents an increase of the CD8+SP TCRhigh thymocytes from 0.5–4% to 30–60% in N15tg/RAG-2−/−/β2M−/− FTOC thymic lobes incubated with 10 μM peptide. For negative selection, the single, open square, the single solid square, and the triple solid squares represent no deletion, 2-fold reduction, and 10-fold reduction, respectively, of the N15tg/RAG-2−/− mice DP thymocytes 24 h after injection of 6-week-old animals with 24 μg peptide i.v. The Kb anchor residues of the peptide are italicized.
Phenotypic Evidence of Positive Selection Induced by the L4 Variant.
To examine the APL effect on positive selection of N15 thymocytes, the N15tg/RAG2−/− (H-2b) were bred onto a β2m−/− background where class I expression is drastically reduced such that these mice lack CD8+ SP cells (33–35). Fetal thymic lobes obtained from N15tg/RAG-2−/−/β2m−/− mice were cultured in the presence of various APLs plus human β2m to restore antigen presentation. As shown in Fig. 1A and summarized in Table 1, only one of the peptides tested, L4, efficiently induces N15 thymocyte maturation as judged by the down-regulation of CD4 (i.e., increase in the percentage and absolute number of CD8+ SP thymocytes) and the up-regulation of the TCR expression level in both DP and SP CD8+ thymocytes (Fig. 1B). As L4 causes reduction in the percentage of DP thymocytes when injected into N15tg/RAG-2−/− mice (Table 1), it may also stimulate a low level of negative selection in FTOC (Fig. 1A). With the exception of VSV8 (Fig. 1A) and I4 (not shown), both of which strongly induce negative selection, all of the other APLs were without effect.
Figure 1.
The L4 peptide induces positive selection of N15tg thymocytes. FTOC was performed by using N15tg/RAG-2−/−/β2m−/− (H-2b) thymic lobes in media containing 5 μg/ml human β2m with or without 10 μM of the indicated peptides. After 7 days, thymocytes were released from the lobes by pressing through a steel mesh, counted, and triple-stained with PE-conjugated anti-CD4, Red613-conjugated anti-CD8, and mAb R53 (anti-N15 β chain clonotype) plus FITC-conjugated anti-rat IgG (19). (A) The CD8 versus CD4 staining profile of total thymocytes is shown. In this representative experiment the yield of CD8+ SP cells after L4 incubation was 13.5 × 104 cells compared with 0.3–2.2 × 104 cells in FTOC incubated in the absence of exogenous peptides or in the presence of the other indicated peptides. The total thymocyte number recovered from VSV8-exposed FTOC was significantly lower (105 cells per lobe) than from FTOC incubated with any of the other peptides (4–6 × 105 cells per lobe). (B) The histograms of the N15 TCRβ chain expression on DP and CD8+ SP thymocytes derived from FTOC incubated with K1 or L4 peptides are shown. Note that the K1 histogram represents data similar to that obtained with the other peptides (except VSV8). The CD8+ SP thymocytes that mature on L4 express a higher level of the TCR than the DP thymocytes harvested from the same lobe. (C) Thymocytes selected on L4 are functionally responsive to VSV8. Thymocytes from the organ cultures described above [cultured with (+) or without (−) L4] or fresh splenocytes from N15tg/RAG-2−/− mouse were assayed for their proliferative response to irradiated EL-4 cells, in the present of rIL-2 and 10 nM VSV8 or 10 μM L4 or no peptide. After 48 h, each well was pulsed for 18 h with [3H]thymidine, harvested on filter discs, and counted. The proliferative responses for the peptides are shown. Results are mean values of triplicate samples with SD noted.
L4-Mediated Functional Maturation.
The above results provide phenotypic evidence of positive selection of fetal N15tg thymocytes in the presence of the L4 peptide. To determine whether the L4 peptide induced functional maturation, thymocytes from N15tg/RAG-2−/−/β2m−/− fetal thymic lobes, which had been cultured in the presence or absence of the L4 peptide, were incubated with antigen-presenting cells pulsed with L4 (10−5 M) or VSV8 (10−8). These peptide concentrations were chosen because ≥1,000-fold higher concentration of the weak agonist L4 was needed to activate mature N15 T cells to a level equivalent to activation by VSV8. Fig. 1C shows that thymocytes positively selected by L4 proliferated in response to VSV8 and, to a lesser extent, to L4. Both sets of responses were significantly greater (P < 0.001) than those of thymocytes that had not matured in the presence of L4. These results argue persuasively for the ability of L4 to induce positive selection of N15 TCR bearing CD8+ SP thymocytes. Experiments performed in an ovalbumin-specific, H-2Kb restricted TCR transgenic model (OVA-TCR) showed that agonist APL cannot mediate positive selection of functional CD8+ T cells (36). In P14 TCR tg mice, the agonist APL induced the appearance of CD8+ SP thymocytes that proliferated to the cognate peptide, and those cells essentially were tolerant to the positively selecting ligands (37). In the N15tg model shown here, the weak agonist ligand (L4) is capable of inducing positive selection of mature SP CD8+ cells that respond to the antigenic peptide as well as the APL that facilitated their differentiation. Hence, the N15 TCR thymocytes are not completely tolerized to the L4 peptide. Interestingly, the antigenic peptide, VSV8, failed to induce positive selection of N15 thymocytes at any concentration tested (10−5 to 10−12 M) (Fig. 1A and data not shown). Negative selection of DP N15 thymocytes could be detected in FTOC incubated with 10−5 to 10−9 M VSV8. At VSV8 concentrations lower than 10−9 M, no changes in the thymocyte number or percentages of DP and SP CD8+ thymocytes could be detected in FTOC. Thus, L4 was the only peptide tested capable of inducing positive selection of N15tg thymocytes.
Crystal Structure of the L4/Kb TCR Ligand.
To begin to understand the differences between the N15 TCR ligands that induced positive selection (L4/Kb) versus negative selection (VSV8/Kb), the crystal structure of L4/Kb was determined, refined to 2.1 Å resolution, and then compared with the VSV8/Kb structure (13, 14). The relevant crystallographic data are summarized in Table 2. The complex crystallized under very similar conditions as the wild-type VSV8/Kb, with both using phosphate as precipitant and a similar pH (7.0 versus 6.25). Both L4/Kb and VSV8/Kb crystals belong to the same space group, P21212, and have similar unit cell dimensions (136.7 Å, 88.0 Å, 45.2 Å for L4/Kb versus 138.1 Å, 88.9 Å, 45.7 Å for VSV8/Kb). The 3% smaller L4/Kb unit cell volume is mostly because of shrinkage resulting from the freezing of the L4/Kb crystal preceding diffraction data collection. Given the overall similarities between these two crystals, it was expected that the two peptide–MHC structures would be very similar, as was indeed the case. Aside from some surface side chains, the only significant changes observed were restricted to the local region around the p4 position of the bound peptide. Although a somewhat different conformation of the Arg at p1 is observed, the lack of clear-cut density for this side chain in the VSV8/Kb structure makes it unlikely to represent a true difference from that of L4/Kb (D. Fremont, personal communication).
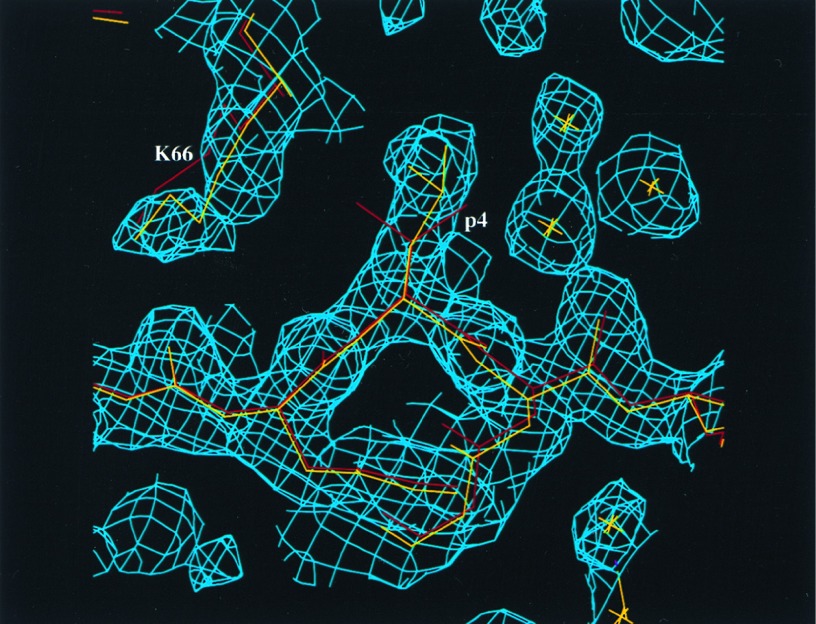
Fig. 2 is a view of a portion of the final 2Fo − Fc difference Fourier with refined VSV8/Kb and L4/Kb structures superimposed on the map. The conformation of the variant peptide’s leucine side chain is unambiguous. In addition, as a consequence of the one methylene group extension from Val-4 in the VSV8 peptide to Leu-4 in the L4 peptide, the exposed side chain of Lys-66 on the α1 helix of the Kb molecule changes its conformation. Two significant polar interactions maintain the ɛ-amino group of this lysyl side chain in the same fixed position. One is a salt bridge (within 2.8 Å) to Glu-63 one helical turn N-terminal to Lys-66 in the α1 domain. The other is a hydrogen bond (within 2.6 Å) to the carbonyl oxygen of the p2 residue of the peptide. Strikingly, the aliphatic portion of the Lys-66 side chain (in particular, the Cγ and Cδ atoms) swings about 2.5 Å inward toward the antigen-binding groove of Kb molecule. Fig. 3 clearly shows that in the VSV8/Kb structure, there is a van der Waals contact between the Cγ2 atom of Val-4 in the VSV8 peptide and the Cδ atom of Lys-66 (3.6 Å apart). In the L4/Kb structure, by comparison, the contact is between the Cδ1 atom of Leu-4 in L4 peptide and Cβ atom of Lys-66 (3.9 Å apart). Apparently, the V4L replacement has caused the conformational change of the Lys-66 side chain. Given that the peptide’s main chain conformation is fixed by p3 and p5 anchoring residues, two tyrosines, and this in turn mandates that the side chain at the p4 position projects against the α1 helical region in the vicinity of the Kb Lys-66 residue, any change in the peptide at p4 likely will affect the conformation of the Lys-66 side chain. VSV8/Kb fails to induce positive selection. Hence, the V4L replacement at p4 plus the attendant conformational change at Lys-66 of Kb most probably are the key structural determinants for positive selection of N15 TCR-bearing DP thymocytes by L4/Kb. The I4 peptide harboring a V4I replacement is functionally indistinguishable from VSV8 (Table 1). It is possible that the Cβ branch present in I4 is critical for maintaining Lys-66 in the “VSV8/Kb conformation.”
Figure 2.
L4/Kb electron density map in the region of the peptide. A portion of the final 2Fo − Fc electron density map calculated with the residue at the p4 position of the peptide omitted. The refined models of both the L4/Kb (yellow) and the VSV8/Kb (red) are superimposed on the map. The map shows how similar the L4/Kb and VSV8/Kb structures are, except for the labeled p4 and Lys-66 side chains of Kb. The figure was prepared by using the program setor (55).
Figure 3.
Comparison of L4/Kb and VSV8/Kb structures reveals the p4 methylene extension and conformational change in the Kb Lys-66 side chain in L4/Kb. Shown here is the schematic representation of the α1+α2 domain of the Kb molecule with side chains of Lys-66 and Glu-63 as well as the two peptides. The color codes are: green for the two Kb helical regions and the side chain of Glu-63; red for the remaining Cα trace of the α1+α2 domains; white for VSV8 peptide and the Lys-66 side chain with which it interacts; and blue for the L4 peptide and the corresponding Lys-66 residue. The figure was prepared with setor (55). Note that the view is such that the TCR would dock roughly from the top of the figure and the Kb α3 and β2m domain would be located at the bottom of the figure, i.e., below this MHC peptide-presenting platform.
General Features of Positive Selection.
What is the nature of positively selecting peptides in general? Depending on the system investigated, it has been reported that peptides with full agonistic, partial agonistic, antagonistic, and null effects on TCR function in mature T lymphocytes all can lead to positive selection of thymocytes (38–42). Affinity measurements by surface plasmon resonance of the interaction between a soluble ovalbumin peptide (SIINFEKL)/Kb-restricted TCR and its peptide ligands suggests that high-affinity ligands are deleting, low-affinity ligands are positively selecting, and functional null ligands are of too low affinity to measure (43). However, measurement of the N15 TCR interaction with VSV8/Kb by similar methodology shows that the monomeric affinity is >200 μM, an affinity 10 times lower than that measured between the OVA/Kb-specific TCR and its positively selecting ligand (unpublished results). Yet VSV8 induces negative selection of N15 TCR-bearing DP thymocytes. These observations suggest that the minimal affinity required for a productive interaction between a TCR and its ligand may vary greatly among different TCRs such that no absolute affinity threshold mandates a positive or negative selection outcome. Nonetheless, one can imagine that the affinity of N15 for L4/Kb may be less than for VSV8/Kb. The postulated alterations in N15 CDR loops required to accommodate the L4/Kb versus VSV8/Kb ligands may occur at a significant energy cost and be associated with a faster off-rate. This difference in biophysics of the interaction may permit activation of genetic programs required for positive selection in the case of the L4/Kb ligand but largely spare activation of the caspase cascade, which leads to cell death during negative selection (21).
Identification of self-peptides extracted from class I MHC molecules on thymic epithelial cell lines shows that peptides that restore positive selection in TCR transgenic TAP-deficient FTOC bear no primary sequence homology to the cognate peptide recognized by that same TCR on mature T lymphocytes (44, 45). Moreover, although the positive-selecting capacity of endogenous peptides is specific for a given TCR, these natural peptides fail to stimulate mature T cells. Furthermore, in genetically engineered mice expressing a thymic MHC class II complex bearing a single peptide, multiple TCRs are selected (46–49). Collectively, these results imply that a positively selecting ligand may bind, but weakly, to the TCRs whose differentiation it triggers. Recognition of such a positively selecting peptide bound to an MHC molecule may involve a subset of elements on the ligand structurally related to but distinct from those of the cognate peptide/MHC complex recognized by the same TCR on mature T cells.
Implications.
Very recently, the crystal structure of N15 TCR in complex with the H57 anti-Cβ Fab fragment has been reported at 2.8 Å resolution as well as a low-resolution structure of the same TCR in complex with VSV8/Kb (7, 15). From these two structures, it is likely that the TCR contact area around the p4-Lys-66 local region consists of Tyr-28, Leu-29, and Ile-30 of the Vα CDR1 loop and possibly Tyr-102 of the Vα CDR3 loop. These are all suitable hydrophobic residues, which might interact with the exposed hydrophobic surface of the local region involving the p4 and Lys-66 side chains. It is tempting to speculate that the focal surface-shape change of this region in L4/Kb versus VSV8/Kb may necessitate corresponding shape changes within the TCR, especially involving those CDR1 loop residues in the Vα domain. This kind of discrete structural plasticity in protein–protein interfaces has been observed in another system (50) and may be the determining factor for the positive selection we have observed experimentally. If CDR1 plays a significant role in this recognition process, as predicted, then positive selection may be encoded, at least in part, in germ-line Vα genes.
This notion would not be unreasonable in view of the fact that the number of endogenous peptides required to generate the T cell repertoire in the thymus must be less than the number of cognate peptides for which various TCRs are specific in the peripheral T cell compartment (42). The preponderance of key residues at peptide positions recognized by the Vα domain also is consistent with the view that the Vα domain has a critical role to play in recognition during thymic development as well as mature T cell function. Moreover, the TCRα subunit is induced late in development relative to the TCRβ subunit, thereby appearing within DP thymocytes in anticipation of subsequent positive selection events. By contrast, β chain expression appears earlier within the DN thymocyte population (51, 52). Conformational alteration of central MHC α-helical residues such as K66 imposed by individual peptide side-chain modifications likely magnifies even small differences among TCR ligands, making the recognition process highly efficient. Previously observed alterations in a given MHC molecule induced by APL variants of a single peptide (53) or unrelated peptides (54) are consistent with this hypothesis. Further mutational and functional studies can clearly test these speculations.
Acknowledgments
This work is supported by National Institutes of Health Grant AI19807 (to E.L.R.), GM56008 (to J.-H.W.), and AI39098 (to H.-C.C.). P.K. was supported by fellowships from the Schweizerischer Nationalfond and Schweizerische Krebsliga. We thank Drs. J. Griffin and L. Clayton for careful review of the manuscript. We also thank Dr. A. Smolyar for helpful comments.
ABBREVIATIONS
- TCR
T cell receptor
- MHC
major histocompatibility complex
- β2m
β2-microglobulin
- DP
double-positive
- SP
single-positive
- FTOC
fetal thymic organ cultures
- APL
altered peptide ligand
- FITC
fluorescein isothiocyanate
Note Added in Proof
We have now determined the crystal structure of I4/Kb. As predicted, the conformation of K66 in I4/Kb structure is similar to that in the VSV8/Kb structure. Both the Cγ1 and Cδ1 atoms of Ile at the p4 position make close contacts (3.4 Å and 2.6 Å, respectively) to the Cδ atom of K66.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID code 10SZ).
References
- 1.Meuer S C, Acuto O, Hercend T, Schlossman S F, Reinherz E L. Annu Rev Immunol. 1984;2:23–50. doi: 10.1146/annurev.iy.02.040184.000323. [DOI] [PubMed] [Google Scholar]
- 2.Marrack P, Kappler J. Adv Immunol. 1986;38:1–30. doi: 10.1016/s0065-2776(08)60005-x. [DOI] [PubMed] [Google Scholar]
- 3.Davis M M, Bjorkman P J. Nature (London) 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 4.Zinkernagel R M, Doherty P C. Nature (London) 1974;248:701–702. doi: 10.1038/248701a0. [DOI] [PubMed] [Google Scholar]
- 5.Garcia K C, Degano M, Stanfield R L, Brunmark A, Jackson M R, Peterson P A, Teyton L, Wilson I A. Science. 1996;274:209–219. [PubMed] [Google Scholar]
- 6.Garboczi D N, Ghosh P, Utz U, Fan Q R, Biddison W E, Wiley D C. Nature (London) 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 7.Wang J-h, Lim K, Smolyar A, Teng M-k, Liu J-h, Tse A G D, Liu J, Hussey R E, Chishti Y, Thomson C T, et al. EMBO J. 1998;17:10–26. doi: 10.1093/emboj/17.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjorkman P J, Saper M A, Samraoui B, Bennett W S, Strominger J L, Wiley D C. Nature (London) 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 9.Saper M A, Bjorkman P J, Wiley D C. J Mol Biol. 1991;219:277–319. doi: 10.1016/0022-2836(91)90567-p. [DOI] [PubMed] [Google Scholar]
- 10.Brown J H, Jardetzky T S, Gorga J C, Stern L J, Urban R G, Strominger J L, Wiley D C. Nature (London) 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 11.Stern L J, Brown J H, Jardetzky T S, Gorga J C, Urban R G, Strominger J L, Wiley D C. Nature (London) 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 12.Fremont D H, Stura E A, Matsumura M, Peterson P A, Wilson I A. Proc Natl Acad Sci USA. 1995;92:2479–2483. doi: 10.1073/pnas.92.7.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fremont D H, Matsumura M, Stura E A, Peterson P A, Wilson I A. Science. 1992;257:919–927. doi: 10.1126/science.1323877. [DOI] [PubMed] [Google Scholar]
- 14.Matsumura M, Fremont D H, Peterson P A, Wilson I A. Science. 1992;257:927–934. doi: 10.1126/science.1323878. [DOI] [PubMed] [Google Scholar]
- 15.Teng M-K, Smolyar A, Tse A G D, Liu J-H, Liu J, Hussey R E, Nathenson S G, Chang H-C, Reinherz E L, Wang J-H. Curr Biol. 1998;8:409–412. doi: 10.1016/s0960-9822(98)70160-5. [DOI] [PubMed] [Google Scholar]
- 16.Garcia K C, Degano M, Pease L R, Huang M, Peterson P A, Teyton L, Wilson I A. Science. 1998;279:1166–1172. doi: 10.1126/science.279.5354.1166. [DOI] [PubMed] [Google Scholar]
- 17.Shibata K-I, Imarai M, van Bleek G M, Joyce S, Nathenson S G. Proc Natl Acad Sci USA. 1992;89:3135–3139. doi: 10.1073/pnas.89.7.3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W, Young A C M, Imarai M, Nathenson S G, Sacchettini J C. Proc Natl Acad Sci USA. 1992;89:8403–8407. doi: 10.1073/pnas.89.17.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang H-C, Smolyar A, Spoerl R, Witte T, Yao Y, Goyarts E C, Nathenson S G, Reinherz E L. J Mol Biol. 1997;271:278–293. doi: 10.1006/jmbi.1997.1169. [DOI] [PubMed] [Google Scholar]
- 20.Ghendler Y, Hussey R E, Witte T, Mizoguchi E, Clayton L K, Bhan A K, Koyasu S, Chang H-C, Reinherz E L. Eur J Immunol. 1997;27:2279–2289. doi: 10.1002/eji.1830270923. [DOI] [PubMed] [Google Scholar]
- 21.Clayton L K, Ghendler Y, Mizoguchi E, Patch R J, Ocain T D, Orth K, Bhan A K, Dixit V M, Reinherz E L. EMBO J. 1997;16:2282–2293. doi: 10.1093/emboj/16.9.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghendler Y, Mizoguchi E, Bhan A K, Clayton L K. Int Immunol. 1998;10:767–774. doi: 10.1093/intimm/10.6.767. [DOI] [PubMed] [Google Scholar]
- 23.Martin S, Bevan M J. Eur J Immunol. 1997;27:2726–2736. doi: 10.1002/eji.1830271037. [DOI] [PubMed] [Google Scholar]
- 24.Chang H-C, Bao Z-Z, Yao Y, Tse A G D, Goyarts E C, Madsen M, Kawasaki E, Brauer P P, Sacchettini J C, Nathenson S G, Reinherz E L. Proc Natl Acad Sci USA. 1994;91:11408–11412. doi: 10.1073/pnas.91.24.11408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 27.Brunger A T. xplor: A System for X-Ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. , Version 3.1. [Google Scholar]
- 28.Evavold B D, Allen P M. Science. 1991;252:1308–1310. [PubMed] [Google Scholar]
- 29.De Magistris M T, Alexander J, Coggeshall M, Altman A, Gaeta F C A, Grey H M, Sette A. Cell. 1992;68:625–634. doi: 10.1016/0092-8674(92)90139-4. [DOI] [PubMed] [Google Scholar]
- 30.Jameson S C, Carbone F R, Bevan M J. J Exp Med. 1993;177:1541–1550. doi: 10.1084/jem.177.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evavold B D, Sloan-Lancaster J, Jeff Wilson K, Rothbard J B, Allen P M. Immunity. 1995;2:655–663. doi: 10.1016/1074-7613(95)90010-1. [DOI] [PubMed] [Google Scholar]
- 32.Janeway C A, Medhzhitov R, Pfeiffer C, Tao X, Bottomly K. The Immunologist. 1995;3/2:41–45. [Google Scholar]
- 33.Zijlstra M, Li E, Sajjadi F, Subramani S, Jaenisch R. Nature (London) 1989;342:435–438. doi: 10.1038/342435a0. [DOI] [PubMed] [Google Scholar]
- 34.Koller B H, Marrack P, Kappler J W, Smithies O. Science. 1990;248:1227–1230. doi: 10.1126/science.2112266. [DOI] [PubMed] [Google Scholar]
- 35.Zijlstra M, Bix M, Simister N E, Loring J M, Raulet D H, Jaenisch R. Nature (London) 1990;344:742–746. doi: 10.1038/344742a0. [DOI] [PubMed] [Google Scholar]
- 36.Hogquist K A, Jameson S C, Bevan M J. Immunity. 1995;3:79–86. doi: 10.1016/1074-7613(95)90160-4. [DOI] [PubMed] [Google Scholar]
- 37.Sebzda E, Kundig T M, Thomson C T, Aoki K, Mak S-Y, Mayer J P, Zamborelli T, Nathenson S G, Ohashi P S. J Exp Med. 1996;183:1093–1104. doi: 10.1084/jem.183.3.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashton R P, Tonegawa S. Immunol Today. 1994;15:362–366. doi: 10.1016/0167-5699(94)90174-0. [DOI] [PubMed] [Google Scholar]
- 39.Jameson S C, Hogquist K A, Bevan M J. Annu Rev Immunol. 1995;13:93–126. doi: 10.1146/annurev.iy.13.040195.000521. [DOI] [PubMed] [Google Scholar]
- 40.Janeway C A. Immunol Today. 1995;16:223–225. doi: 10.1016/0167-5699(95)80163-4. [DOI] [PubMed] [Google Scholar]
- 41.Nakano N, Rooke R, Benoist C, Mathis D. Science. 1997;275:678–683. doi: 10.1126/science.275.5300.678. [DOI] [PubMed] [Google Scholar]
- 42.Bevan M J. Immunity. 1997;7:175–178. doi: 10.1016/s1074-7613(00)80520-8. [DOI] [PubMed] [Google Scholar]
- 43.Alam S M, Travers P J, Wung J L, Nasholds W, Redpath S, Jameson S C, Gascoigne N R J. Nature (London) 1996;381:616–620. doi: 10.1038/381616a0. [DOI] [PubMed] [Google Scholar]
- 44.Hogquist K A, Tomlinson A J, Kieper W C, McGargill M A, Hart M C, Naylor S, Jameson S C. Immunity. 1997;6:389–399. doi: 10.1016/s1074-7613(00)80282-4. [DOI] [PubMed] [Google Scholar]
- 45.Hu Q, Bazemore Walker C R, Girao C, Opferman J T, Sun J, Shabanowitz J, Hunt D F, Ashton-Rickardt P G. Immunity. 1997;7:221–231. doi: 10.1016/s1074-7613(00)80525-7. [DOI] [PubMed] [Google Scholar]
- 46.Liu C-P, Parker D, Kappler J, Marrack P. J Exp Med. 1997;186:1441–1450. doi: 10.1084/jem.186.9.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ignatowicz L, Rees W, Pacholczyk R, Ignatowicz H, Kushnir E, Kappler J, Marrack P. Immunity. 1997;7:179–186. doi: 10.1016/s1074-7613(00)80521-x. [DOI] [PubMed] [Google Scholar]
- 48.Tourne S, Miyazaki T, Oxenius A, Klein L, Fehr T, Kyewski B, Benoist C, Mathis D. Immunity. 1997;7:187–195. doi: 10.1016/s1074-7613(00)80522-1. [DOI] [PubMed] [Google Scholar]
- 49.Sure C D, Lee D-S, Fung-leung W-p, Karlsson L, Sprent J. Immunity. 1997;7:209–219. doi: 10.1016/s1074-7613(00)80524-5. [DOI] [PubMed] [Google Scholar]
- 50.Atwell S, Ultsch M, De Vos A M, Wells J A. Science. 1997;278:1125–1128. doi: 10.1126/science.278.5340.1125. [DOI] [PubMed] [Google Scholar]
- 51.von Boehmer H. Cell. 1994;76:219–228. doi: 10.1016/0092-8674(94)90330-1. [DOI] [PubMed] [Google Scholar]
- 52.Shinkai Y, Koyasu S, Nakayama K-i, Murphy K M, Loh D Y, Reinherz E L, Alt F W. Science. 1993;259:822–825. doi: 10.1126/science.8430336. [DOI] [PubMed] [Google Scholar]
- 53.Reid S W, McAdam S, Smith K J, Klenerman P, O’Callaghan C A, Harlos K, Jakobsen B K, McMichael A J, Bell J I, Stuart D I, Jones E Y. J Exp Med. 1996;184:2279–2286. doi: 10.1084/jem.184.6.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Madden D R, Garboczi D N, Wiley D C. Cell. 1993;75:693–708. doi: 10.1016/0092-8674(93)90490-h. [DOI] [PubMed] [Google Scholar]
- 55.Evans S V. J Mol Graphics. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]