Abstract
Adenoviruses (Ads) have evolved multiple mechanisms to evade the host immune response. Several of the immunomodulatory Ad proteins are encoded in early transcription unit 3 (E3). The E3/19K protein interferes with antigen presentation and T cell recognition, whereas the E3/10.4K, 14.5K, and 14.7K proteins can protect cells from tumor necrosis factor α-mediated lysis. Here, we describe an additional activity of E3 proteins. Transfectants expressing all E3 proteins of Ad2 exhibit a profound reduction of the apoptosis receptor CD95 (Fas, APO-1) on the cell surface. In contrast, cells expressing only the E3A region have normal Fas levels. Thus, one of the E3B proteins (10.4K, 14.5K, or 14.7K) seems to be responsible for this effect. To identify the E3B products involved, each individual E3B ORF was selectively disrupted. Examination of stable cell lines containing the mutated E3 regions showed that Fas expression is restored when either the 10.4K or the 14.5K ORF is disrupted, whereas mutation of the 14.7K ORF does not rescue Fas expression. Loss of Fas on the cell surface is accompanied by a similar decrease of total Fas levels. However, in the presence of lysosomotropic agents Fas accumulates in endosomal/lysosomal vesicles, indicating that 10.4K–14.5K induce internalization and degradation of Fas. Down-regulation of Fas but not CD40 is also observed during infection and as a consequence, Ad-infected cells are protected from Fas-mediated apoptosis. Thus, the Fas system is implicated in Ad pathogenesis.
Keywords: CD95/down-regulation/immune evasion
Apoptosis or programmed cell death is considered as an effective antiviral defense mechanism of the host (1, 2). Virus-infected cells undergo apoptosis in response to signals originating from two basically distinct cellular locations: (i) The signal may originate endogenously, for example, from unscheduled DNA synthesis, induction of the cell cycle or disruption of the cellular metabolism (2). (ii) Apoptosis of infected cells is also triggered by exogenous stimuli mostly produced by the host immune system. Examples are cytolysis by cytotoxic T cells, natural killer cells, and tumor necrosis factor (TNF) α (3, 4). As a common principle, a soluble or membrane-bound ligand produced by the immune cell interacts with an apoptosis receptor of the TNF receptor/nerve growth factor receptor (TNFR/NGFR) family.
Fas (APO-1 or CD95) belongs to the TNFR/NGFR family and is constitutively expressed on a variety of tissues (5). Expression of the Fas ligand is restricted to some immune privileged sites, natural killer cells, and is induced in activated T cells upon recognition of antigenic peptide/major histocompatibility complexes on target cells (5). The latter mechanism is thought to be important for down-regulation of an immune response but it also allows cytotoxic T lymphocytes to induce Fas-mediated apoptosis (3–5).
Fas ligand binding induces the oligomerization of Fas and the recruitment of adaptor molecules such as FADD (MORT). This in turn triggers the association and activation of the cysteine protease caspase 8 (FLICE, Mach, Mch5) that activates other caspases (6) ultimately leading to apoptosis (5, 6). Viral interference with premature apoptosis of infected cells is a prerequisite for effective reproduction of viruses and may be important for establishing viral persistence (1, 2). Viral anti-apoptotic proteins have been described that target various steps of the apoptosis cascade. Several have homology to cellular Bcl-2 family members, others inhibit the apoptosis mediator p53 (1, 2) or interfere with signal transduction from the cell surface receptor (2, 6, 7).
Human adenoviruses (Ads) can cause acute as well as persistent infections (8). A key role for regulating the interaction of the virus with its host and presumably for persistence has been attributed to proteins encoded in the non essential early transcription unit 3 (E3) of the virus (9, 10). We previously showed that the E3/19K protein blocks transport of class I major histocompatibility antigens to the cell surface (11), thereby interfering with antigen presentation and T cell recognition (10, 12). The E3/14.7K, 10.4K, and 14.5K proteins can protect cells from TNF-mediated lysis (9). Moreover, TNF stimulates expression of E3 proteins by activating the transcription factor NF-κB that binds to and activates the E3 promoter (13, 14). Additional in vivo data using animal models for Ad induced disease (15) or E3 transgenic mice further support the idea that E3 proteins modulate the immune response (16, 17).
We now report that the E3/10.4K–14.5K proteins down-regulate Fas from the cell surface, by inducing its internalization and degradation in endosomes or lysosomes. As a consequence, Ad-infected cells are protected from Fas-mediated apoptosis. Infection with Ad2 does not affect CD40, another member of the TNFR/NGFR family, suggesting a rather selective interference with these type of cell death receptors. The data indicate that Fas and its ligand are important effector molecules in the anti-adenoviral immune response.
MATERIALS AND METHODS
Construction of Mutant Genes.
Plasmid pBS-E3 containing the EcoRV C fragment of Ad2 in pBluescript II KS (Stratagene) was cleaved with KpnI and ClaI to delete the XhoI site in the multiple cloning site. After blunting and religation the resulting plasmid pBSΔX-E3 was cleaved at the XhoI site within the 10.4K gene. Blunting and religation generated plasmid pBSΔX-E3–10.4* containing a 4-base frameshift 13 bp downstream of the E3/10.4K start codon. Expression of the E3/14.5K gene was disrupted by mutating the ATG to AACG thereby eliminating the start codon and introducing a 1-base frame shift and a new PacI site. PCR-mediated mutagenesis was done as described (18), using the two mutant oligonucleotides CTTTAATTAACGAAACGGAGTGTC and CCGTTTCGTTAATTAAAGAATTCTG and ATTGGACGGTCTGAAACC and GCTTGACCACACAAAAGATACC located 5′ and 3′, respectively, of the side to be mutagenized (all oligos are given in 5′ to 3′ orientation, mutated nucleotides are underlined). The mutagenized PCR fragment was cleaved by XhoI and HpaI, and the 773-bp fragment was ligated into pBSΔX-E3 generating pBSΔX-E3–14.5*. Similarly, a 1-base frameshift was inserted into the E3/14.7K gene 18 bp downstream of the E3/14.7K start codon disrupting a BglII site by using GAATCTCTAGACTCTAGAATTGG and CCAATTCTAGAGTCTAGAGATTCAGTC as mutant primers and ATTGGACGGTCTGAAACC and GGAATTTGACATCCC as flanking primers. The XhoI–HpaI fragment was cloned into pBSΔX-E3 giving rise to pBSΔX-E3–14.7*. All constructs were verified by dye terminator cycle sequencing.
Transfection, Cell Lines, Culture Conditions, and Viruses.
Transfection of human 293 cells with the modified E3 plasmids and the pSV2-neor plasmid was done as described (18). 293E3–45 and 293E3A-22 cells are derivatives of 293 cells obtained by transfection with the EcoRV C and EcoRI D fragments of Ad2, respectively (13).
CD40 expressing HeLa cells were kindly provided by H. Engelmann (19), and MCF-Fas cells and derivatives MCF-FasBcl2 and MCF-FasVI generated by supertransfection of MCF-Fas with a Bcl-2 expression vector or the empty vector alone were kindly provided by M. Jäättelä (20). Cells were cultured as described (18).
Ad2 and H2dl801 viruses were propagated and titrated on A549 cells by plaque assay or a fluorescence-activated cell sorter (FACS)-based assay staining for hexon with mAb 2Hx-2.
mAbs and Antisera.
The following mAbs were used: Tw1.3 (21), against E3/19K; 2Hx-2, ATCC HB-8117, anti-hexon of Ad2; B-G27, (Dianova, Hamburg, Germany), anti-APO-1 (22), CH-11 (PharMingen) against human CD95; W6/32, ATCC HB-95, anti-HLA-ABC; 528, ATCC HB-8509, anti-epidermal growth factor receptor (EGFR); 5E9C11, ATCC HB-21, anti-transferrin receptor; G28–5 (23), anti-CD40; 2D5, anti-Lamp-2, (24); SJ1D1, anti-CD13; J4–48, anti-CD46 (Coulter Immunotech).
Polyclonal antisera were raised against the synthetic peptides CYRDRTIADLLRIL, CEISYFNLTGGDD, and CGIRDLIPFN, corresponding to the C termini of the E3/10.4K, E3/14.5K, and E3/14.7K proteins, respectively. Coupling and immunization was performed as described (18). Rabbit serum against Fas was purchased from Santa Cruz Biotechnology or was provided by H. Engelmann.
Flow Cytometry.
FACS analysis was carried out essentially as described (18). In some experiments (Fig. 4 A–C) the cells were fixed with formaldehyde (CellFIX, Becton Dickinson) before staining.
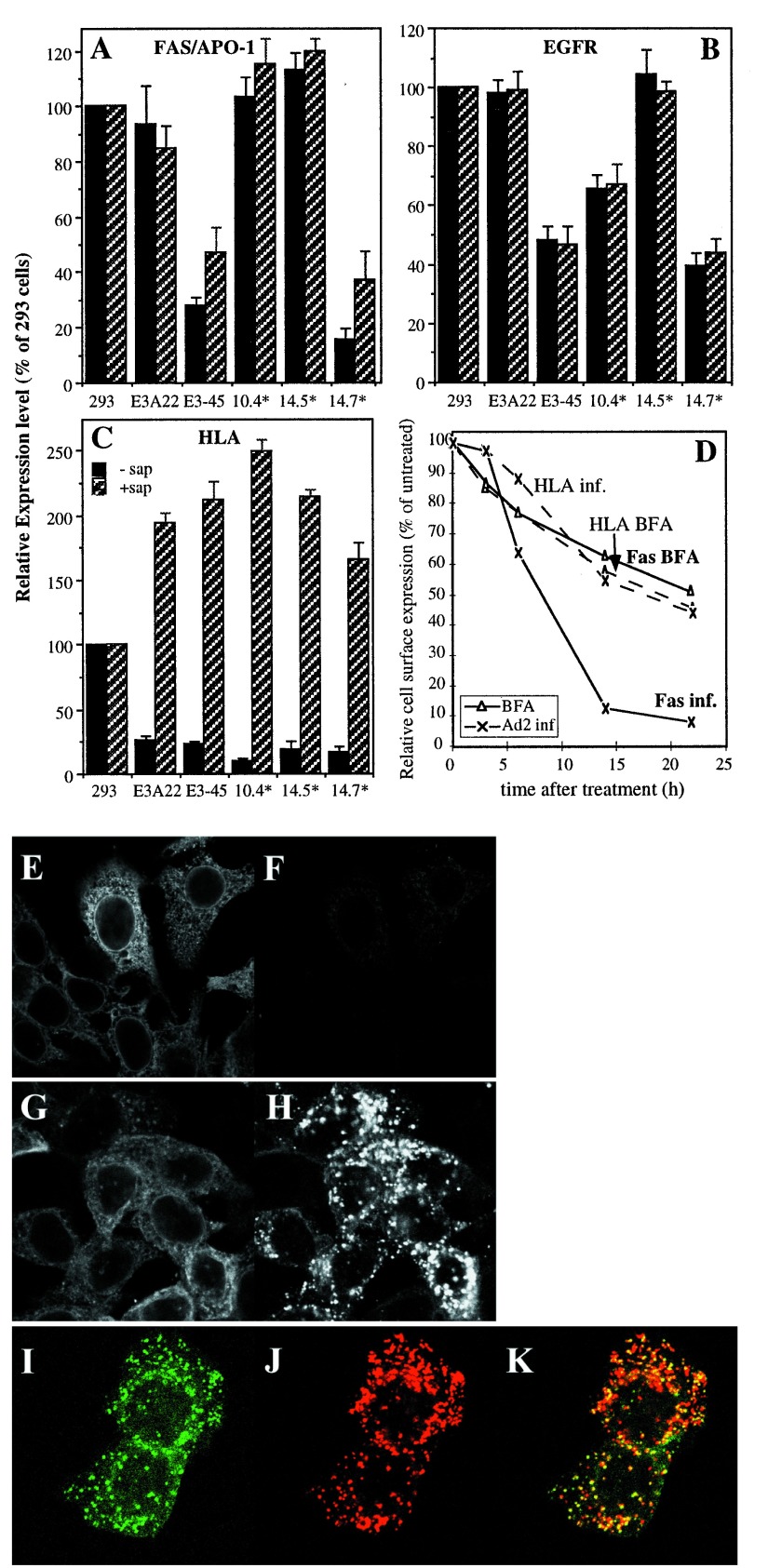
Figure 4.
Ad E3 proteins trigger endocytosis and intracellular degradation of Fas. (A–C) Relative expression of plasma membrane proteins as detected by FACS analysis in the presence or absence of 0.1% saponin. Before incubation with the antibodies against HLA (C, W6/32), Fas (A, B-G27) and EGFR (B, 528), cells were fixed and processed as described in Materials and Methods. After deduction of the values obtained with an IgG2a isotype control for B-G27 the mean value of fluorescence for each cell line was related to that of 293 cells determined in the same experiment that was set to 100%. The data are compiled from at least five independent experiments using 293, 293E3A–22, 293E3–45 and the three mutant clones mentioned above. Analysis of other clones gave similar results (data not shown). The black bars denote the values obtained in the absence, the hatched bars those in the presence of saponin. Error bars represent the SEM. (D) A549 cells were either infected with Ad2 or incubated with 5 μg/ml brefeldin A (BFA). After 3, 6, 14, and 22 h, cell surface expression of HLA and Fas was analyzed by FACS. (E–K) Intracellular localization of Fas by laser scanning confocal microscopy. Cells were infected with Ad2. 6 h postinfection cells were mock treated (E and F) or incubated with 100 nM bafilomycin A1 (G and H) or 50 mM NH4Cl (I–K) for further 9 h. After fixation and permeabilization cells were stained with E3/19K specific mAb Tw1.3 (E and G), a rabbit antiserum against Fas (F, H, and I) or mAb 2D5 against human Lamp-2 (J). The overlay of I and J is shown in K. A significant number of vesicles are yellow indicating colocalization of Fas and Lamp-2.
Cell Labeling, Immunoprecipitation, SDS/PAGE, and Immunoblotting.
Metabolic labeling of cells, immunoprecipitation, and SDS/PAGE were carried out essentially as described (11). Western blot analysis was as described (18) except that a Trans-Blot SD semi-dry transfer cell (Bio-Rad) was used and specific bands were detected by using horseradish peroxidase conjugated antibodies (1:1000) against rabbit IgG (Dianova) and 4-chloro-1-naphthol/H2O2.
MTT Tetrazolium Assay.
The amount of cell death is the reciprocal value of the viability as determined by the MTT assay (25). Briefly, ≈20,000 cells per well were plated in 96-well microtiter plates in 100 μl of complete medium. Twenty-four hours later cells were infected or mock treated. Seven hours postinfection 10 μl of arabinofuranosyl cytosine (final concentration: 20 μg/ml) was added and 5 h later half of the cultures were supplemented with anti-CD95 antibody CH-11 (200–500 ng/ml). After further incubation for 48 h at 37°C, cells received 1/10 volume of MTT (5 mg/ml, Sigma). Three hours later cells were centrifuged and lysed by addition of 150 μl of acidic isopropyl alcohol (70% isopropyl alcohol, 0.03 M HCl). Blue formazan crystals were dissolved by vigorous pipetting and incubation at 4°C for 2–12 h. The absorption at 570 nm was determined with a microplate reader using a reference wavelength of 620 nm.
Immunofluorescence.
Cells were processed for immunofluorescence as described (11) with the following modification, all antibody incubations (40 min) were performed in PBS containing 0.2% saponin and 5% fetal calf serum. Cells were examined with a laser scanning confocal microscope (Leitz DM IRB; scanner, Leica TCS 4D).
RESULTS
E3B Products Interfere with Cell Surface Expression of Fas.
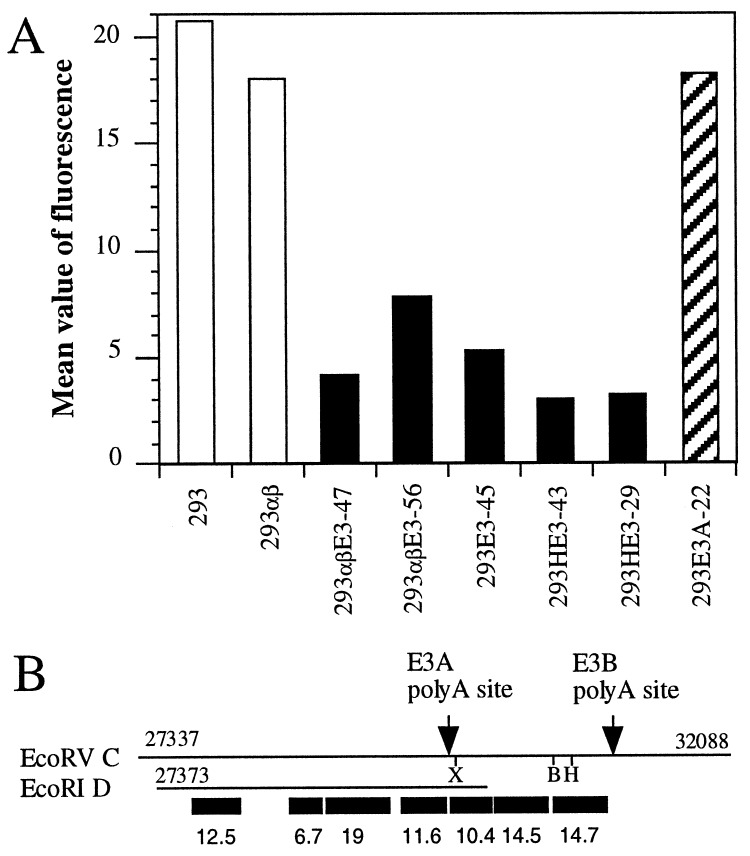
By monitoring the expression of cell surface molecules on E3+ 293 cells we noted that all randomly chosen 293 transfectants derived from independent transfections with the entire E3 region of Ad2 exhibit a drastic reduction of the apoptosis receptor CD95 (also named APO-1 or Fas; Fig. 1A, ▪) on the cell surface whereas 293 cells express reasonable amounts of Fas. In contrast, 293E3A-22 transfectants containing the EcoRI D fragment and allowing only expression of the 12.5K, 6.7K, 19K, and 11.6K proteins (Fig. 1B) appear to have normal levels of Fas (Fig. 1A). Similar results were obtained for other independently derived E3A transfectants (data not shown). Thus, we conclude that an Ad protein encoded in the E3B region is responsible for down-regulation of Fas.
Figure 1.
Down-regulation of Fas cell surface expression in E3+ 293 cells. 293 cells and derivatives were stained for cell surface expression of Fas using the mAb anti-APO-1. Flow cytometry revealed on average an 80% reduction of Fas cell surface expression on E3+ 293 cells (■). 293αβ cells expressing human major histocompatibility complex class II genes served as control transfectants. H indicates cotransfection with the hygromycin resistance gene. The data represent the mean of two experiments. Background staining obtained with the secondary antibody alone was deducted. (B) Schematic representation of the E3 transcription unit of Ad2. The two lines refer to the two DNA fragments used to generate the above transfectants. The EcoRV C fragment includes all E3 genes whereas the EcoRI D fragment only encompasses 12.5K, 6.7K, 19K, 11.6K, the E3A poly(A) site and a truncated 10.4K gene. As this DNA fragment lacks the E3B poly(A) site 10.4K is not expressed. Poly(A) sites and the restriction sites used for mutating and cloning are indicated. X, XhoI; B, BglII; H, HpaI. ORFs, ■.
Selective Inactivation of E3B Products.
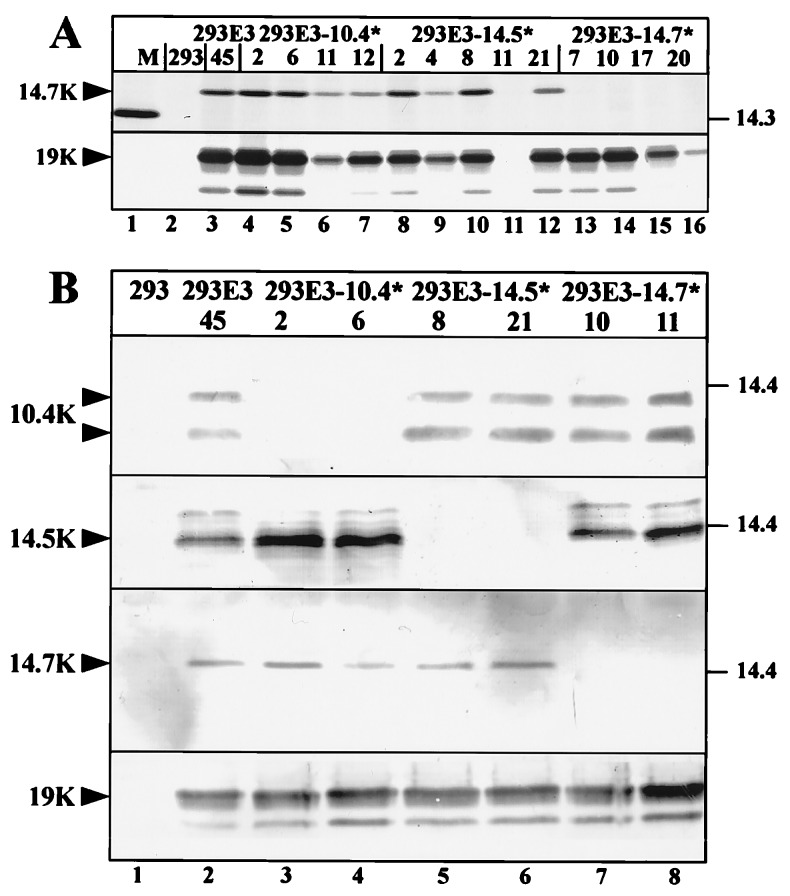
The E3B region is highly conserved among human Ads and codes for three proteins called 10.4K, 14.5K, and 14.7K. To identify which of these proteins is required for the observed down-regulation of Fas, we selectively disrupted each of the three E3B ORFs by mutating the ATG start codon and/or by introducing a frame shift. Mutagenesis was designed such to keep alterations to a minimum (1 and 4 bp insertions) because it was previously shown that deletions within this region profoundly affect splicing, thereby increasing 10.4/14.5K expression and severely reducing expression of other E3 proteins, in particular E3/19K (9, 26). Incorporating these mutations into the complete E3 region plasmid, the E3/10.4K*, E3/14.5K* and the E3/14.7K* plasmids were generated in which expression of the 10.4K, 14.5K and the 14.7K proteins, respectively, was expected to be lost. By transfection into 293 cells permanent cell clones were established. Four clones from each transfection expressing similar amounts of E3/19K as 293E3–45 cells, as judged by FACS analysis in the presence of the detergent saponin (data not shown), were chosen for further analysis. Immunoprecipitation of E3/19K confirmed that the expression level of E3/19K in these clones is comparable to that of 293E3–45 cells (Fig. 2A Lower, 19K). The 14.7K specific antiserum precipitates a protein of ≈16 kDa whose expression level correlates with that of the 19K protein in 293E3–45, 293E3–10.4* and 293E3–14.5* transfectants (Fig. 2A, lanes 3–12), showing that the introduced mutations did not affect the expression of neighboring genes. In contrast, no 14.7K protein was detected in cells containing the mutated 14.7K gene (Fig. 2A, lanes 13–16). Thus, expression of the 14.7K product seems to be selectively eliminated.
Figure 2.
Selective knock-out of E3B protein expression. (A) Immunoprecipitation of E3/14.7K and E3/19K from 293 cells (lane 2), 293E3–45 cells containing the wild-type Ad2 E3 transcription unit (lane 3), 293E3–10.4* cell clones (lanes 4–7), 293E3–14.5* cell clones (lanes 8–12) and 293E3–14.7* cell clones (lanes 13–16). 5 × 106 cells were labeled with 100 μCi/ml (1 Ci = 37 GBq) of each [35S]methionine and [35S]cysteine for 3 h, 1 ml detergent lysates were immunoprecipitated with mAb Tw1.3 against the E3/19K protein or with a rabbit antiserum against the E3/14.7K protein. Immunoprecipitates were subjected to SDS/PAGE on an 11.5% to 13.5% gradient gel. The migration positions for the 14.7K and the 19K proteins are indicated by arrowheads. Molecular masses of the marker proteins (M) are given in kDa. 293E3–14.5*-11 is a G418-resistant clone that does not express E3 proteins. (B) Western blot analysis of mutant cell lines. Immunoprecipitates of detergent extracts from similar number of cells (1.5 × 107) were separated by SDS/PAGE and analyzed by western blotting with the same antisera used for immunoprecipitation. E3/19K was immunoprecipitated with mAb Tw1.3 and probed with an E3/19K specific rabbit serum (18). Specific bands are indicated.
The expression of the 14.5K and 10.4K proteins in the transfectants was followed by combining immunoprecipitation with Western blot analysis. In 293E3–45 cells and transfectants containing mutant E3/14.5K and E3/14.7K genes (Fig. 2B, lanes 2, 5–8) but not in transfectants containing mutant E3/10.4K genes (lanes 3 and 4) the 10.4K specific antiserum labels two protein species of ≈12 kDa and 14 kDa, which seem to correspond to the two described isoforms of the 10.4K protein with and without signal sequence (9, 27). Similarly, the 14.5K products are visualized in all E3 transfectants except in those containing mutant E3/14.5K genes (Fig. 2B, lanes 2–8). Detection of the 19K and 14.7K proteins by Western blot analysis corroborated the FACS and immunoprecipitation data (Fig. 2A). Taken together, we conclude that the introduced mutation affects selectively the expression of the mutated gene and thus the transfectants can be utilized to identify the Fas regulating E3 protein.
E3/10.4K and 14.5K Proteins Are Responsible for Fas Repression.
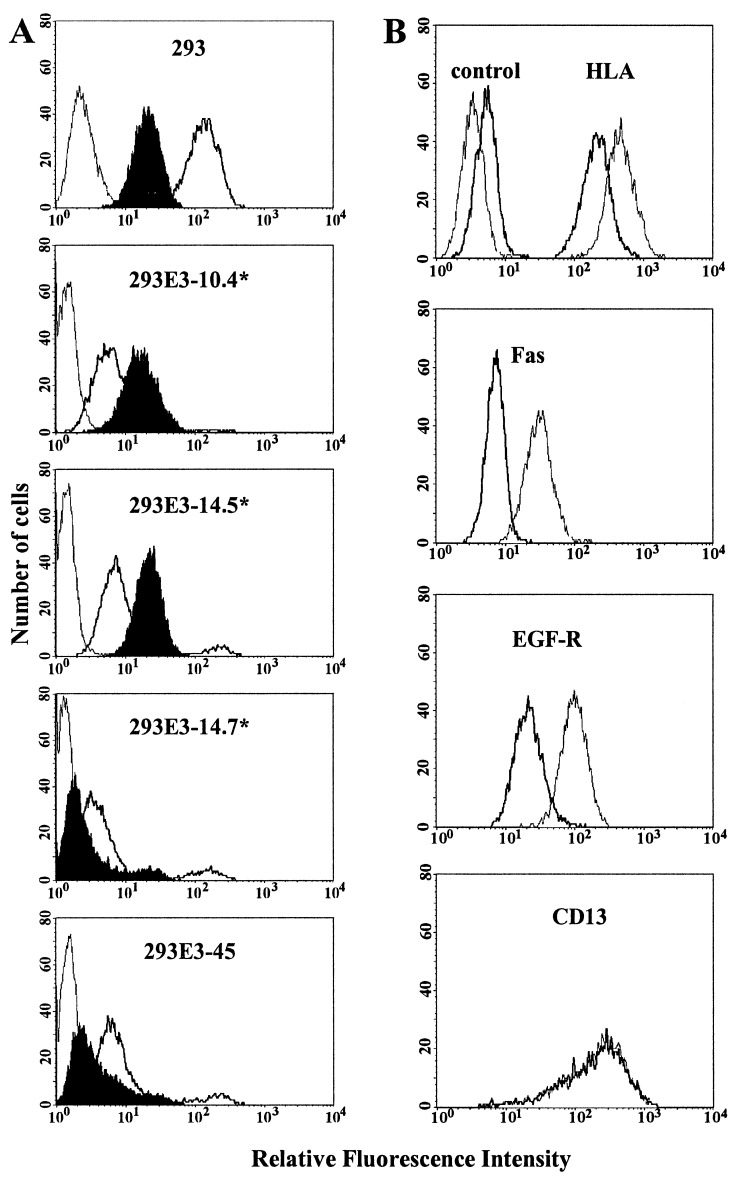
Next we tested the various transfectants for cell surface expression of Fas. 293E3–14.7* mutant cells like the 293E3–45 cells exhibited drastically reduced levels of Fas, whereas cell surface expression was restored in both 293E3–10.4* and 293E3–14.5* cells (Figs. 3A and 4A), indicating that both E3/10.4K and 14.5K proteins are required for Fas suppression. This is consistent with previous reports indicating that E3/10.4K and 14.5K proteins form a physical complex (9, 28). Cell surface expression of the EGFR is also significantly decreased (Fig. 4B, EGFR). However, down-regulation is selective because neither the transferrin receptor nor CD46 expression is specifically influenced by E3 products (data not shown). Down-regulation of HLA in all E3 transfectants reflects the activity of the E3/19K protein (Fig. 4C, HLA, ▪).
Figure 3.
(A) Cell surface expression of Fas is restored in 293 transfectants with mutant 10.4K and 14.5K ORFs. Cells were stained with FITC-labeled goat anti-mouse IgG (thin line), the HLA specific mAb W6/32 (bold line) and the Fas specific mAb B-G27 (filled histogram). At least five independent experiments with at least three clones from each transfection were performed. In this representative experiment the mutant cell clones 293E3–10.4*-2, 293E3–14.5*-8 and 293E3–14.7*-10 were analyzed. (B) Modulation of cell surface proteins upon infection with Ad2. A549 cells were infected with 50 multiplicity of infection per cell of Ad2 for 32 h or were mock treated. The cell surface proteins analyzed are given in the histogram. As control, cells were incubated with fluorescein isothiocyanate-labeled goat anti-mouse IgG. Staining of mock-treated cells is indicated by thin lines, that of infected cells by bold lines.
Down-Regulation of Fas Is Also Observed during Infection with Ad2.
Fig. 3B shows that down-modulation of Fas is not an isolated property of 293 transfectants but can also be observed upon infection of the human lung epitheloid cell line A549. A549 cells were mock treated (Fig. 3B, plane line) or infected with Ad2 for 32 h (Fig. 3B , bold line) and analyzed by FACS by using mAbs against HLA, Fas/APO-1, the EGFR, and CD13. Cell surface expression of HLA is reduced upon infection by 40% whereas that of CD95 reaches almost background levels (Fig. 3B, compare Fas and control histograms). A profound reduction is also observed for the EGFR but not for CD13. Similar results were obtained for HeLa cells, the human breast carcinoma cell line MCF-Fas or normal human diploid lung (MRC-5) and foreskin fibroblasts SeBu (data not shown). Down-regulation of Fas is also observed upon infection with Ad5 but not with mutant viruses H5dl327 or H2dl801 containing deletions within the E3 region of Ad5 and Ad2, respectively. Thus, E3 Proteins affect Fas expression in transformed and normal cells.
Fate of Fas in E3+ Cells.
To follow the fate of Fas molecules in E3+ cells FACS analysis was performed in the presence of the mild detergent saponin allowing the additional detection of intracellular proteins. In contrast to the intracellular accumulation of HLA observed in the presence of E3/19K (Fig. 4C), the disappearance of Fas on the cell surface is accompanied by a comparable decrease of intracellular/total Fas (Fig. 4A, compare ▪ with ▨). Similarly, the removal of the EGFR from the cell surface is paralleled by a comparable intracellular loss (Fig. 4B, ▨), although there are some differences: The relative loss of intracellular/total Fas was less pronounced and differed by 20% compared with that on the cell surface, whereas the relative loss of the EGFR in the presence or absence of saponin is basically identical. Also, a differential sensitivity of Fas and the EGFR to the 14.5K product is observed (Fig. 4A, B, compare 10.4* with 293E3–45). No E3-dependent reduction was observed for the transferrin receptor or CD46 (data not shown).
The comparable intracellular loss suggests that the 10.4K–14.5K proteins may down-regulate Fas by a mechanism similar or identical to that previously suggested for down-regulation of the EGFR by these proteins (9, 29), involving internalization and degradation in endosomes/lysosomes. RT-PCR did not reveal significant differences in Fas RNA levels between E3+ and E3− cells (data not shown). To clarify whether or not Fas is endocytosed upon Ad2 infection, MCF-Fas cells overexpressing Fas were infected with Ad2 or were mock-treated. 15 h postinfection cells were prepared for laser scanning confocal microscopy. In accord with the FACS data, the typical rim around the cells indicative for cell surface staining was lost upon infection, but in general no significant intracellular Fas staining was observed in cells expressing E3/19K that was used as a marker for infected cells (Fig. 4 E and F). Only when infected cells were treated with lysosomotropic agents such as Bafilomycin A1, an inhibitor of the endosomal ATPase, ammonium chloride or chloroquine Fas accumulates in intracellular vesicles (Fig. 4 G and H, and data not shown). A significant proportion of these also stains for the lysosomal membrane protein 2 (Lamp-2; Fig. 4 I–K). This indicates that E3/10.4–14.5K redirect Fas into the endosomal/lysosomal system where it is degraded.
Two pathways have been described for entering this system, one from the trans-Golgi network and a second one from the cell surface via early and late endosomes (30, 31). To distinguish at which location Fas is rerouted, we compared the decline of cell surface Fas after infection with that seen after BFA treatment (Fig. 4D). If the E3 proteins redirect Fas transport in the trans-Golgi network, no Fas molecule should reach the cell surface anymore and thus the outcome should be similar to that upon BFA treatment that also inhibits export out of the Golgi. As predicted from the known transport inhibitory effect of E3/19K on HLA, the slow decay of HLA seen after infection parallels that observed BFA treatment. In contrast, Fas disappears much more rapidly upon infection than it does upon BFA treatment, suggesting that E3/10.4–14.5K act on cell surface Fas.
Adenovirus Infection Protects Cells from Fas-Mediated Apoptosis.
We next tested whether E3-mediated down-regulation of Fas is functionally significant and interferes with Fas-mediated apoptosis. To this end, the Fas overexpressing cell lines MCF-Fas, MCF-FasVI, and MCF-FasBcl2, a double transfectant overexpressing also Bcl-2, were mock treated or infected with Ad2 or a mutant virus H2dl801 containing a deletion of all E3 genes except 12.5 and 14.7 (32, 33). Twelve to 16 h after infection, half of the cultures received mAb CH-11 that crosslinks and activates Fas. Viability was monitored 48 h later with the MTT assay. Compared with mock treated cells, Fas-induced apoptosis was profoundly reduced when cells were infected with Ad2 (Table 1). This appears to be mainly due to the activity of E3 proteins because the mutant virus H2dl801 reduced Fas-induced apoptosis only little. Even in cells overexpressing Bcl-2, which are largely resistant to Fas-mediated apoptosis (20), the small amount of Fas induced cell death can be prevented by infection with Ad2. Thus, Ad2 harbors E3 gene products that can protect cells from Fas-mediated apoptosis.
Table 1.
Inhibition of Fas-mediated cell death upon infection with Ad2
| Cell line | Mock | Ad2 | H2dl801 |
|---|---|---|---|
| MCF-Fas | 55.2 ± 1.4 | 11.0 ± 3.9 | 43.1 ± 3.3 |
| MCF-FasVI | 52.9 ± 2.7 | 2.2 ± 6.3 | 36.6 ± 3.3 |
| MCF-FasBcl2 | 25.4 ± 2.9 | 5.8 ± 3.8 | 12.5 ± 6.4 |
Viability of cells in the absence and presence of Fas antibodies was determined in triplicates by the MTT assay and the percentage of Fas-mediated cell death was calculated relative to that of untreated cells (see Materials and Methods). Data represent the mean ± SEM of five experiments.
Adenovirus Does Not Significantly Affect Cell Surface Expression of CD40.
To gain insight into the specificity of the above reported E3 effect, we examined the sensitivity of CD40, another member of the TNFR/NGFR superfamily. CD40 is expressed on B cells and plays a central role in T cell-dependent antibody responses. Ligation of CD40 with its cognate ligand CD40L expressed on T cells initiates a sequence of events leading to B cell activation (34). In addition, triggering of CD40 on transformed cells can induce apoptosis (19). Recently, it was reported that transient subversion of CD40L function diminishes immune responses to adenovirus gene therapy vectors and prolongs their persistence (34, 35). Therefore, it was interesting to study the effect of E3 expression on CD40. Examination of CD40 overexpressing HeLa cells (19) after infection with Ad2 by FACS analysis did not reveal a significant loss of CD40 surface staining compared with mock treated cells (Fig. 5). In contrast, during the same time Fas expression was almost completely abolished. Similar results were obtained for the squamous cervix carcinoma cell line SiHa that expresses under the control of the endogenous promoter ≈1/6 of the amount of CD40 compared with HeLaCD40 (data not shown). Thus, CD40 appears not to be affected by expression of the 10.4K–14.5K complex.
Figure 5.
Expression of CD40 in uninfected and Ad2 infected HeLa-CD40 cells. HeLa-CD40 cells (19) were infected with 50 multiplicity of infection of Ad2 for 24 h (solid histogram) or were mock treated (bold line). Flow cytometry was performed with mAbs G28–5 and B-G27 directed to human CD40 and Fas, respectively. Staining of mock and infected cells with the control secondary reagents is indicated with thin and dotted lines, respectively.
DISCUSSION
Here we show that the adenovirus E3 proteins 10.4K–14.5K efficiently down-regulate cell surface expression of the apoptosis receptor CD95. The effect is observed in established E3+ cell lines upon transfection (Figs. 1 and 3) as well as during infection of human lung epithelial cells A549 (Fig. 3B), human cervix carcinoma cells (HeLa, Fig. 5), breast carcinoma cells (MCF-Fas, data not shown) and normal human diploid foreskin and lung fibroblasts (SeBu, MRC-5, data not shown). This indicates that the effect is neither cell type or tissue specific, nor does it require immortalization. Thus, the phenomenon is likely to be relevant for the virus life cycle in vivo.
Selective disruption of the 10.4K, 14.5K, and 14.7K ORF demonstrated that both the 10.4K and 14.5K proteins are required for removal of Fas from the cell surface (Figs. 2–4). Similar conclusions were drawn by using mutant viruses with deletions in E3 (36). Using the same viral mapping system the 10.4K and 14.5K proteins have previously been shown to be responsible for down-regulation of the EGFR (9, 28), although conflicting data suggest that 10.4K alone may be sufficient for this activity (27, 29, 37). The reasons for these obvious discrepancies are not immediately apparent but mapping E3 gene functions with the use of mutant viruses containing deletions in E3 is complicated by the fact that these deletions often affect splicing and therefore the expression of neighboring genes (9, 26, 36). A further complication is that the mutant viruses are based on a recombinant Ad2/Ad5 virus expressing 10.4K from Ad2 and 14.5K of Ad5. Our approach utilizing E3+ cell lines with selective disruptions of single E3 ORFs of Ad2 did not reveal significant changes in the expression level of E3 products encoded by neighboring E3 genes (Fig. 2). Furthermore, we confirm the importance of 14.5K for down-regulation of the EGFR whereas the presence of 10.4K did not suffice for this effect (Fig. 4B).
Coimmunoprecipation of 10.4K with 14.5K and their apparent mutual interdependence for cell surface expression and inhibition of TNF-mediated lysis suggest that these proteins act as a complex (9, 28). As previously proposed for the down-regulation of the EGFR (9, 29, 37), we suggest that a complex of 10.4K and 14.5K binds to Fas on the cell surface and induces its internalization into endosomes/lysosomes where it is degraded (Fig. 4). This proposed mechanism is consistent with the presence of LL and YxxΦ sorting motifs in the cytoplasmic tails of 10.4K and 14.5K, respectively. Both sequence motifs have been implicated in targeting proteins to the endosomal/lysosomal compartments (30, 31). In agreement with the proposed mechanism, we find concomitant to the loss of Fas on the cell surface of E3/10.4–14.5K+ cells a comparable decrease in intracellular/total Fas levels (Fig. 4A). The relative reduction of staining in the presence of saponin is not as pronounced as that on the cell surface, which is consistent with the idea that degradation of endocytosed Fas is somewhat delayed. This is not the case for the EGFR, which appears to be degraded immediately. This differential behavior could be due to a differential sensitivity to intracellular proteases, a difference in the rate of delivery or may indicate distinct trafficking pathways. In support of the latter, the level of EGFR on the cell surface of 10.4* knock-out cells is only partially increased compared with 10.4K–14.5K+ cells whereas that of Fas is completely restored (Fig. 4 A and B). Analysis of a second 10.4* clone confirmed the differential sensitivity of the EGFR and Fas toward 14.5K (expression of the EGFR, 83%; Fas, 117.3%). Thus, in the absence of 10.4K, 14.5K alone is able to affect expression of the EGFR but not that of the Fas receptor. It is possible, that the 10.4K and 14.5K proteins are unable to bind individually to Fas or a Fas adaptor molecule, although this may occur, at least to some extent, with the EGFR. Alternatively, the individual E3 protein might not be transported to the Fas containing compartment, but still reaches the EGFR. Together, the data indicate that 10.4K–14.5K use similar but not identical mechanisms to down-modulate Fas and the EGFR.
At present, it is unclear whether 10.4K–14.5K directly interact with Fas and the EGFR or the interaction is mediated by a common adaptor molecule associated with these receptors. There is neither an obvious structural similarity between Fas and the EGFR, nor was a common adaptor protein identified so far. To gain insight into the structural requirements for 10.4K–14.5K mediated down-regulation the sensitivity for this effect of CD40, another member of the TNFR/NGFR family that is thought to play an important role for anti-adenoviral immunity (35) was examined. No significant down-regulation of CD40 was observed during Ad2 infection of HeLa or SiHa cells (Fig. 5, and data not shown), yet at the same time Fas expression was almost completely abolished (reduction, 95%). This differential sensitivity could be explained by a differential turnover of the two molecules on the cell surface (Fas high, CD40 low). However, it is well known that the turnover of Fas is slow compared with other proteins. Also, the insensitivity of CD40 seems not to be due to a limited capacity of 10.4K–14.5K to remove the ≈15-fold higher amounts of CD40 as compared with Fas, because CD40 cell surface expression is also not diminished in SiHa cells exhibiting CD40 levels comparable to the Fas levels of MCF-Fas cells. Therefore, Fas/CD40 chimeras should allow us to identify the structural domains within Fas that sensitize this molecule for 10.4–14.5K mediated down-modulation.
Overall the presence of Fas modulating adenovirus proteins suggests that Fas and Fas ligand are important host effector molecules that the virus tries to counteract early during infection. Whether their importance relates to a Fas based killing mechanism of Ad specific cytotoxic T lymphocytes and/or natural killer cells or stems from an as yet unknown anti-viral host response remains to be shown.
Acknowledgments
We are grateful to H. Engelmann, M. Jäättelä, S. Hess, A. Hasilik, and P. Krammer for the gifts of antibodies and cells. Special thanks to S. B. for providing SeBu cells. We thank W. Muranyi for advice with the confocal microscope and C. Ebenau-Jehle and A. Osterlehner for excellent technical assistance. For critical reading of the manuscript and helpful comments we thank H. Engelmann, S. Grimm, M. Windheim, and A. Iglesias. This work was supported by grants SFB 388 and Bu 642/1 (to H.-G.B.) of the German Science Foundation.
ABBREVIATIONS
- Ad2
adenovirus type 2
- BFA
brefeldin A
- CTL
cytotoxic T cells
- EGFR
epidermal growth factor receptor
- E3
early transcription unit 3
- TNF
tumor necrosis factor
- TNFR/NGFR
TNF receptor/nerve growth factor receptor
- FACS
fluorescence-activated cell sorter
Note
. After submission of this manuscript results analogous to those reported in Figs. 3B and 4 E–K and Table 1 were published (38).
References
- 1.Shen Y, Shenk T E. Curr Opin Genet Dev. 1995;5:105–111. doi: 10.1016/s0959-437x(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 2.Teodoro J G, Branton P E. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berke G. Cell. 1995;81:9–12. doi: 10.1016/0092-8674(95)90365-8. [DOI] [PubMed] [Google Scholar]
- 4.Nagata S, Golstein P. Science. 1995;267:1449–1456. doi: 10.1126/science.7533326. [DOI] [PubMed] [Google Scholar]
- 5.Nagata S. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 6.Salvesen G S, Dixit V M. Cell. 1997;91:443–446. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- 7.Bertin J, Armstrong R C, Ottilie S, Martin D A, Wang Y, Banks S, Wang G H, Senkevich T G, Alnemri E S, Moss B, et al. Proc Natl Acad Sci USA. 1997;94:1172–1176. doi: 10.1073/pnas.94.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horwitz M S. In: Field’s Virology. Fields B N, Knipe D M, Howley P M, et al., editors. Philadelphia: Lippincott-Raven; 1996. pp. 2149–2171. [Google Scholar]
- 9.Wold W S M, Hermiston T W, Tollefson A E. Curr Top Microbiol Immunol. 1995;199/I:237–274. doi: 10.1007/978-3-642-79496-4_13. [DOI] [PubMed] [Google Scholar]
- 10.Burgert H-G. Trends Microbiol. 1996;4:107–112. doi: 10.1016/0966-842X(96)81527-7. [DOI] [PubMed] [Google Scholar]
- 11.Burgert H-G, Kvist S. Cell. 1985;41:987–997. doi: 10.1016/s0092-8674(85)80079-9. [DOI] [PubMed] [Google Scholar]
- 12.Burgert H-G, Maryanski J L, Kvist S. Proc Natl Acad Sci USA. 1987;84:1356–1360. doi: 10.1073/pnas.84.5.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Körner H, Fritzsche U, Burgert H-G. Proc Natl Acad Sci USA. 1992;89:11857–11861. doi: 10.1073/pnas.89.24.11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deryckere F, Burgert H-G. J Biol Chem. 1996;271:30249–30255. doi: 10.1074/jbc.271.47.30249. [DOI] [PubMed] [Google Scholar]
- 15.Ginsberg H S, Lundholm Beauchamp U, Horswood R L, Pernis B, Wold W S, Chanock R M, Prince G A. Proc Natl Acad Sci USA. 1989;86:3823–3827. doi: 10.1073/pnas.86.10.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Efrat S, Fejer G, Brownlee M, Horwitz M S. Proc Natl Acad Sci USA. 1995;92:6947–6951. doi: 10.1073/pnas.92.15.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Herrath M G, Efrat S, Oldstone M B, Horwitz M S. Proc Natl Acad Sci USA. 1997;94:9808–9813. doi: 10.1073/pnas.94.18.9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sester M, Burgert H-G. J Virol. 1994;68:5423–5432. doi: 10.1128/jvi.68.9.5423-5432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hess S, Engelmann H. J Exp Med. 1996;183:159–167. doi: 10.1084/jem.183.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jäättelä M, Benedict M, Tewari M, Shayman J A, Dixit V M. Oncogene. 1995;10:2297–2305. [PubMed] [Google Scholar]
- 21.Cox J H, Bennink J R, Yewdell J W. J Exp Med. 1991;174:1629–1637. doi: 10.1084/jem.174.6.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trauth B C, Klas C, Peters A M, Matzku S, Moller P, Falk W, Debatin K M, Krammer P H. Science. 1989;245:301–305. doi: 10.1126/science.2787530. [DOI] [PubMed] [Google Scholar]
- 23.Clark E A, Ledbetter J A. Proc Natl Acad Sci USA. 1986;83:4494–4498. [PubMed] [Google Scholar]
- 24.Diettrich O, Gallert F, Hasilik A. Eur J Cell Biol. 1996;69:99–106. [PubMed] [Google Scholar]
- 25.Mosmann T. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 26.Brady H A, Wold W S. Mol Cell Biol. 1988;8:3291–3297. doi: 10.1128/mcb.8.8.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffman P, Yaffe M B, Hoffman B L, Yei S, Wold W S, Carlin C. J Biol Chem. 1992;267:13480–13487. [PubMed] [Google Scholar]
- 28.Tollefson A E, Stewart A R, Yei S P, Saha S K, Wold W S. J Virol. 1991;65:3095–3105. doi: 10.1128/jvi.65.6.3095-3105.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffman P, Carlin C. Mol Cell Biol. 1994;14:3695–3706. doi: 10.1128/mcb.14.6.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trowbridge I S, Collawn J F, Hopkins C R. Annu Rev Cell Biol. 1993;9:129–161. doi: 10.1146/annurev.cb.09.110193.001021. [DOI] [PubMed] [Google Scholar]
- 31.Marks M S, Ohno H, Kirchhausen T, Bonifacino J S. Trends Cell Biol. 1997;7:124–128. doi: 10.1016/S0962-8924(96)10057-X. [DOI] [PubMed] [Google Scholar]
- 32.Challberg S S, Ketner G. Virology. 1981;114:196–209. doi: 10.1016/0042-6822(81)90265-8. [DOI] [PubMed] [Google Scholar]
- 33.Wang E W, Scott M O, Ricciardi R P. J Virol. 1988;62:1456–1459. doi: 10.1128/jvi.62.4.1456-1459.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grewal I S, Borrow P, Pamer E G, Oldstone M B A, Flavell R A. Curr Opin Immunol. 1997;9:491–497. doi: 10.1016/s0952-7915(97)80100-8. [DOI] [PubMed] [Google Scholar]
- 35.Yang Y, Su Q, Grewal I S, Schilz R, Flavell R A, Wilson J M. J Virol. 1996;70:6370–6377. doi: 10.1128/jvi.70.9.6370-6377.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shisler J, Yang C, Walter B, Ware C F, Gooding L R. J Virol. 1997;71:8299–8306. doi: 10.1128/jvi.71.11.8299-8306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carlin C R, Tollefson A E, Brady H A, Hoffman B L, Wold W S. Cell. 1989;57:135–144. doi: 10.1016/0092-8674(89)90179-7. [DOI] [PubMed] [Google Scholar]
- 38.Tollefson A, Hermiston T W, Lichtenstein D L, Colle C F, Tripp P A, Dimitrov T, Toth K, Wells C E, Doherty P C, Wold W S M. Nature (London) 1998;392:726–729. doi: 10.1038/33712. [DOI] [PubMed] [Google Scholar]