Abstract
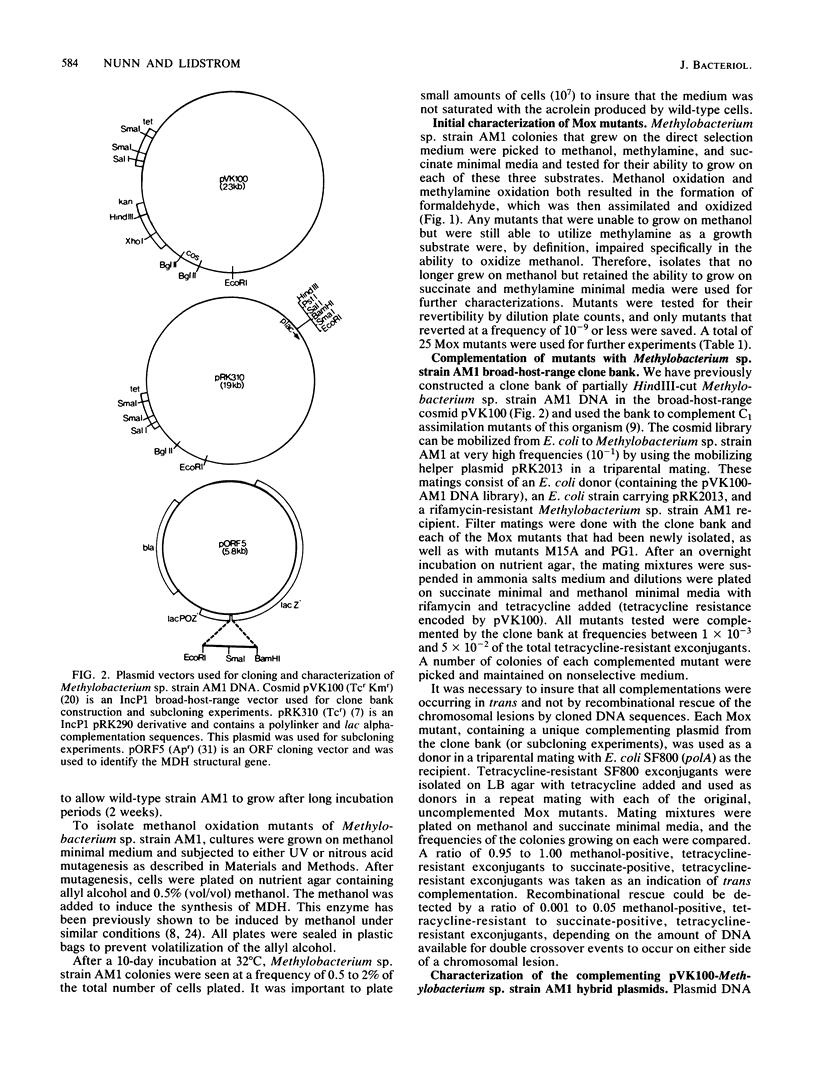
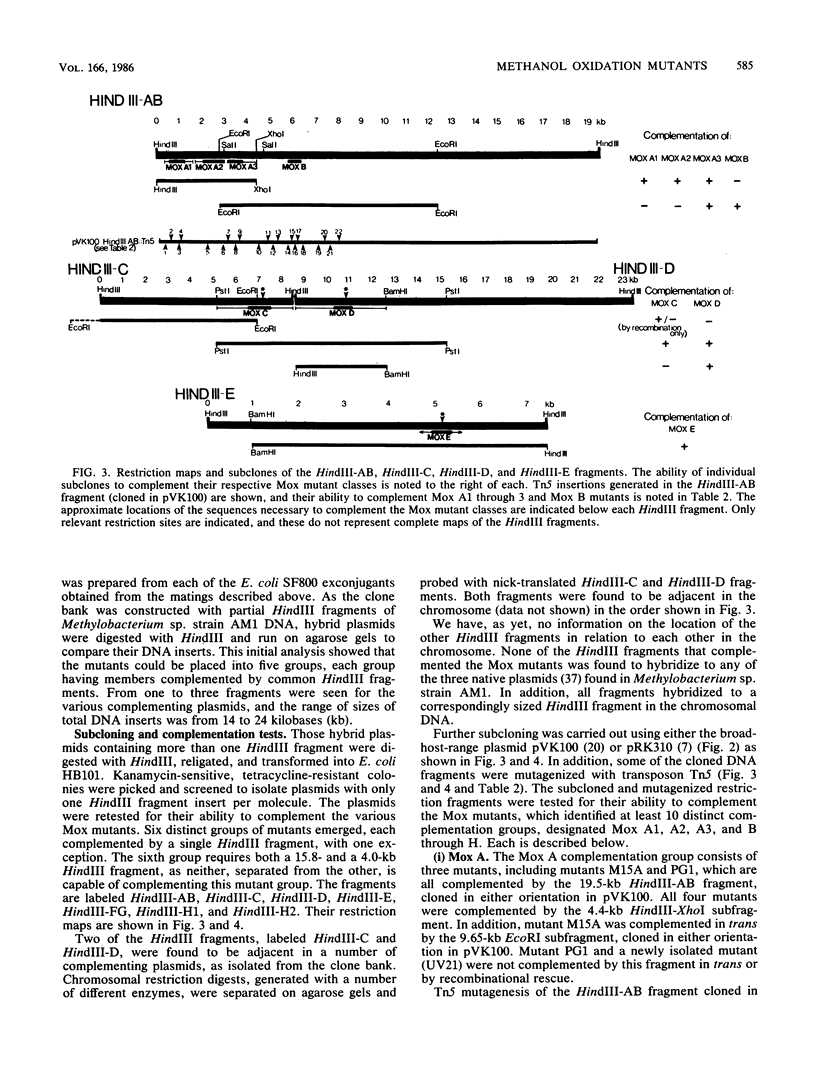
A method has been developed for the direct selection of methanol oxidation mutants of the facultative methylotroph Methylobacterium sp. strain AM1 (formerly Pseudomonas sp. strain AM1). Using this direct selection technique, we have isolated mutants of Methylobacterium sp. strain AM1 that are no longer capable of growth on methanol but retain the ability to grow on methylamine. These methanol oxidation (Mox) mutants were complemented with a genomic clone bank of this organism constructed in the broad-host-range cosmid pVK100, and subcloning and Tn5 mutagenesis experiments have assigned the Mox mutants to 10 distinct complementation groups. Using an open reading frame beta-galactosidase fusion vector and antibodies specific for Methylobacterium sp. strain AM1 methanol dehydrogenase, we have identified the methanol dehydrogenase structural gene and determined the direction of transcription. The results suggest that the synthesis and utilization of an active methanol dehydrogenase in this organism requires at least 10 different gene functions.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen L. N., Hanson R. S. Construction of broad-host-range cosmid cloning vectors: identification of genes necessary for growth of Methylobacterium organophilum on methanol. J Bacteriol. 1985 Mar;161(3):955–962. doi: 10.1128/jb.161.3.955-962.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony C., Zatman L. J. The microbial oxidation of methanol. The alcohol dehydrogenase of Pseudomonas sp. M27. Biochem J. 1965 Sep;96(3):808–812. doi: 10.1042/bj0960808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Chory J., Kaplan S. The in vitro transcription-translation of DNA and RNA templates by extracts of Rhodopseudomonas sphaeroides. Optimization and comparison of template specificity with Escherichia coli extracts and in vivo synthesis. J Biol Chem. 1982 Dec 25;257(24):15110–15121. [PubMed] [Google Scholar]
- Ditta G., Schmidhauser T., Yakobson E., Lu P., Liang X. W., Finlay D. R., Guiney D., Helinski D. R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985 Mar;13(2):149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- Dunstan P. M., Anthony C., Drabble W. T. Microbial metabolism of C 1 and C 2 compounds. The role of glyoxylate, glycollate and acetate in the growth of Pseudomonas AM1 on ethanol and on C 1 compounds. Biochem J. 1972 Jun;128(1):107–115. doi: 10.1042/bj1280107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton G. L., Nunn D. N., Lidstrom M. E. Molecular cloning of a malyl coenzyme A lyase gene from Pseudomonas sp. strain AM1, a facultative methylotroph. J Bacteriol. 1984 Nov;160(2):718–723. doi: 10.1128/jb.160.2.718-723.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel D. J., Simpson R. B., Ream L. W., White F. F., Gordon M. P., Nester E. W. Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell. 1981 Nov;27(1 Pt 2):143–153. doi: 10.1016/0092-8674(81)90368-8. [DOI] [PubMed] [Google Scholar]
- Gautier F., Bonewald R. The use of plasmid R1162 and derivatives for gene cloning in the methanol-utilizing Pseudomonas AM1. Mol Gen Genet. 1980;178(2):375–380. doi: 10.1007/BF00270487. [DOI] [PubMed] [Google Scholar]
- Gershoni J. M., Palade G. E. Protein blotting: principles and applications. Anal Biochem. 1983 May;131(1):1–15. doi: 10.1016/0003-2697(83)90128-8. [DOI] [PubMed] [Google Scholar]
- Hager D. A., Burgess R. R. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980 Nov 15;109(1):76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- Heffron F., Bedinger P., Champoux J. J., Falkow S. Deletions affecting the transposition of an antibiotic resistance gene. Proc Natl Acad Sci U S A. 1977 Feb;74(2):702–706. doi: 10.1073/pnas.74.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Feramisco J. R., Fiddes J. C., Thomas G. P., Hughes S. H. Identification of clones that encode chicken tropomyosin by direct immunological screening of a cDNA expression library. Proc Natl Acad Sci U S A. 1983 Jan;80(1):31–35. doi: 10.1073/pnas.80.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heptinstall J., Quayle J. R. Pathways leading to and from serine during growth of Pseudomonas AM1 on C1 compounds or succinate. Biochem J. 1970 Apr;117(3):563–572. doi: 10.1042/bj1170563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauf V. C., Nester E. W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982 Jul;8(1):45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lorowitz W., Clark D. Escherichia coli mutants with a temperature-sensitive alcohol dehydrogenase. J Bacteriol. 1982 Nov;152(2):935–938. doi: 10.1128/jb.152.2.935-938.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEGNET R. ALKOHOLDEHYDROGENASEMUTANTEN VON SCHIZOSACCHAROMYCES POMBE. Pathol Microbiol (Basel) 1965;28:50–57. [PubMed] [Google Scholar]
- McNerney T., O'connor M. L. Regulation of enzymes associated with C-1 metabolism in three facultative methylotrophs. Appl Environ Microbiol. 1980 Aug;40(2):370–375. doi: 10.1128/aem.40.2.370-375.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinkoth J., Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984 May 1;138(2):267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- Miller L., Kaplan S. Plasmid transfer and expression in Rhodopseudomonas sphaeroides. Arch Biochem Biophys. 1978 Apr 15;187(1):229–234. doi: 10.1016/0003-9861(78)90028-0. [DOI] [PubMed] [Google Scholar]
- PEEL D., QUAYLE J. R. Microbial growth on C1 compounds. I. Isolation and characterization of Pseudomonas AM 1. Biochem J. 1961 Dec;81:465–469. doi: 10.1042/bj0810465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G. B., Ausubel F. M. A general method for site-directed mutagenesis in prokaryotes. Nature. 1981 Jan 1;289(5793):85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- Shultz J., Silhavy T. J., Berman M. L., Fiil N., Emr S. D. A previously unidentified gene in the spc operon of Escherichia coli K12 specifies a component of the protein export machinery. Cell. 1982 Nov;31(1):227–235. doi: 10.1016/0092-8674(82)90422-6. [DOI] [PubMed] [Google Scholar]
- Steers E., Jr, Cuatrecasas P., Pollard H. B. The purification of beta-galactosidase from Escherichia coli by affinity chromatography. J Biol Chem. 1971 Jan 10;246(1):196–200. [PubMed] [Google Scholar]
- Toukdarian A. E., Lidstrom M. E. Molecular construction and characterization of nif mutants of the obligate methanotroph Methylosinus sp. strain 6. J Bacteriol. 1984 Mar;157(3):979–983. doi: 10.1128/jb.157.3.979-983.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]





