Abstract
Mutant P1 Ap Cm lysogens were isolated in which the drug resistance genes resident on the plasmid prophage P1 Ap Cm are amplified by a novel mechanism. The first step required for amplification is IS1-mediated rearrangement of the P1 Ap Cm prophage. The drug resistance genes are amplified from the rearranged P1 Ap Cm prophage by the formation of a plasmid (P1dR) which contains the two resistance genes. The P1dR plasmid is an independent replicon about one-half the size of P1 Ap Cm that can be maintained at a copy number eightfold higher than that at which P1 Ap Cm can be maintained. It contains no previously identified replication origin and is dependent on the Rec+ function of the host.
Full text
PDF

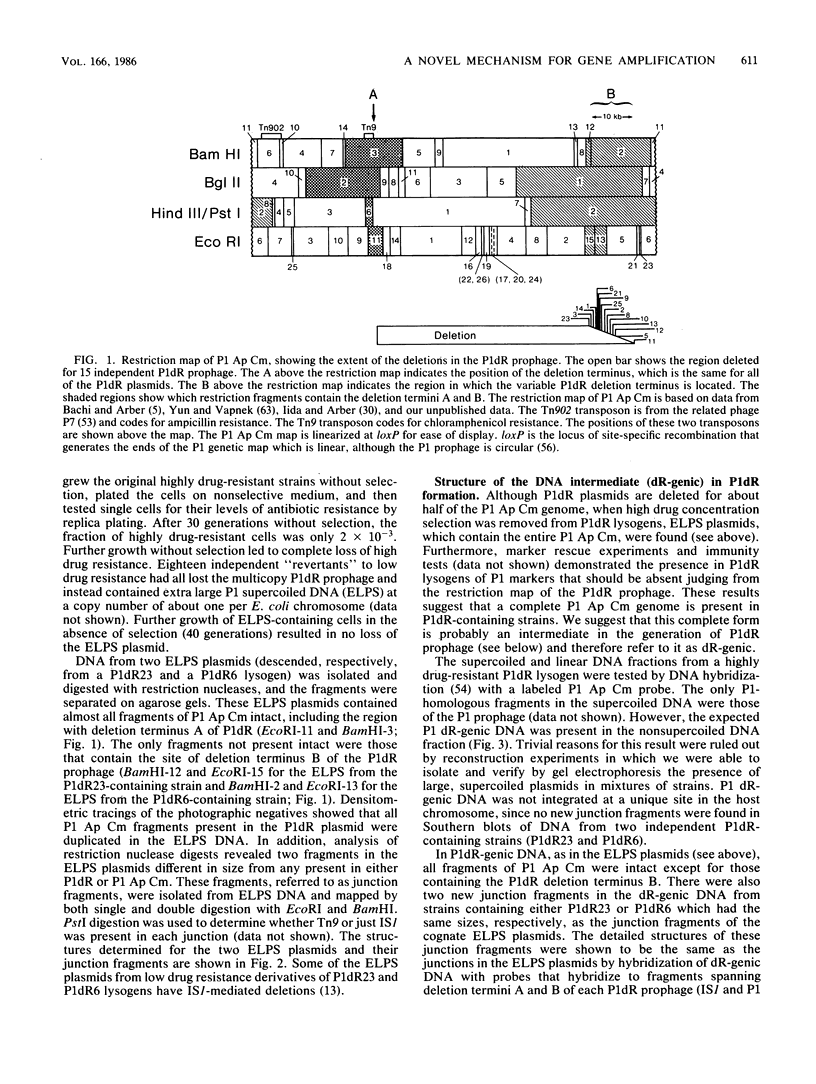
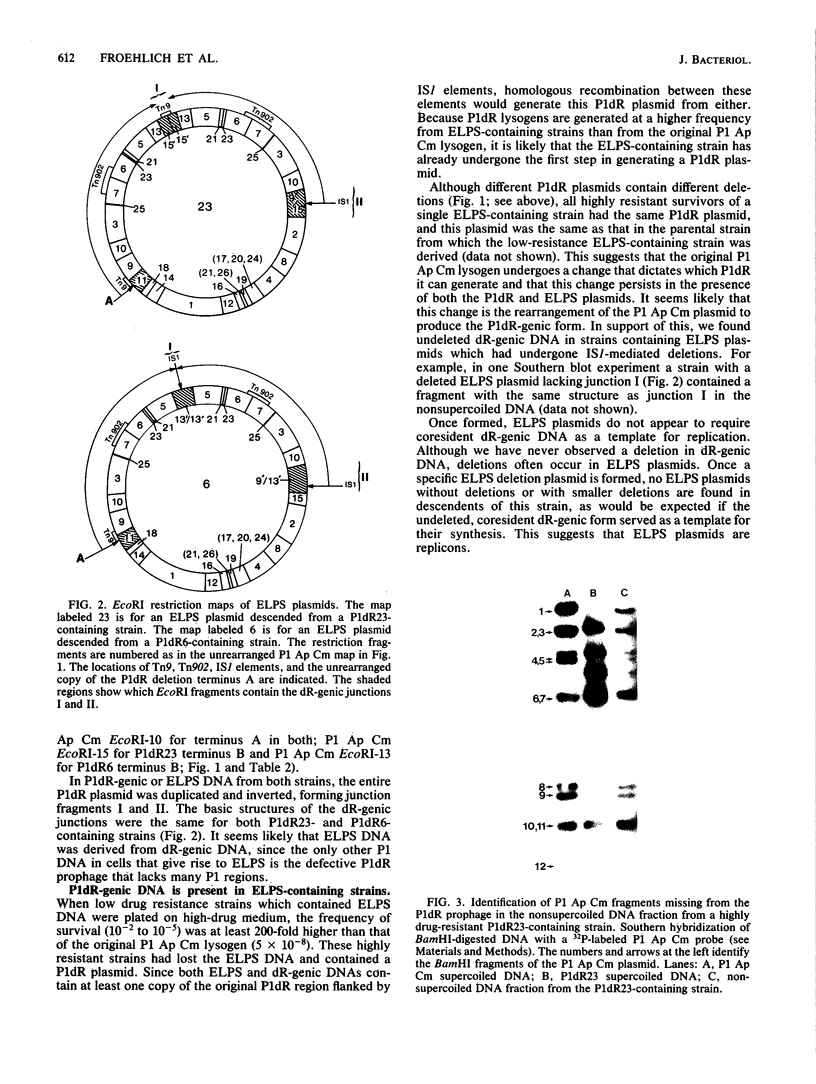
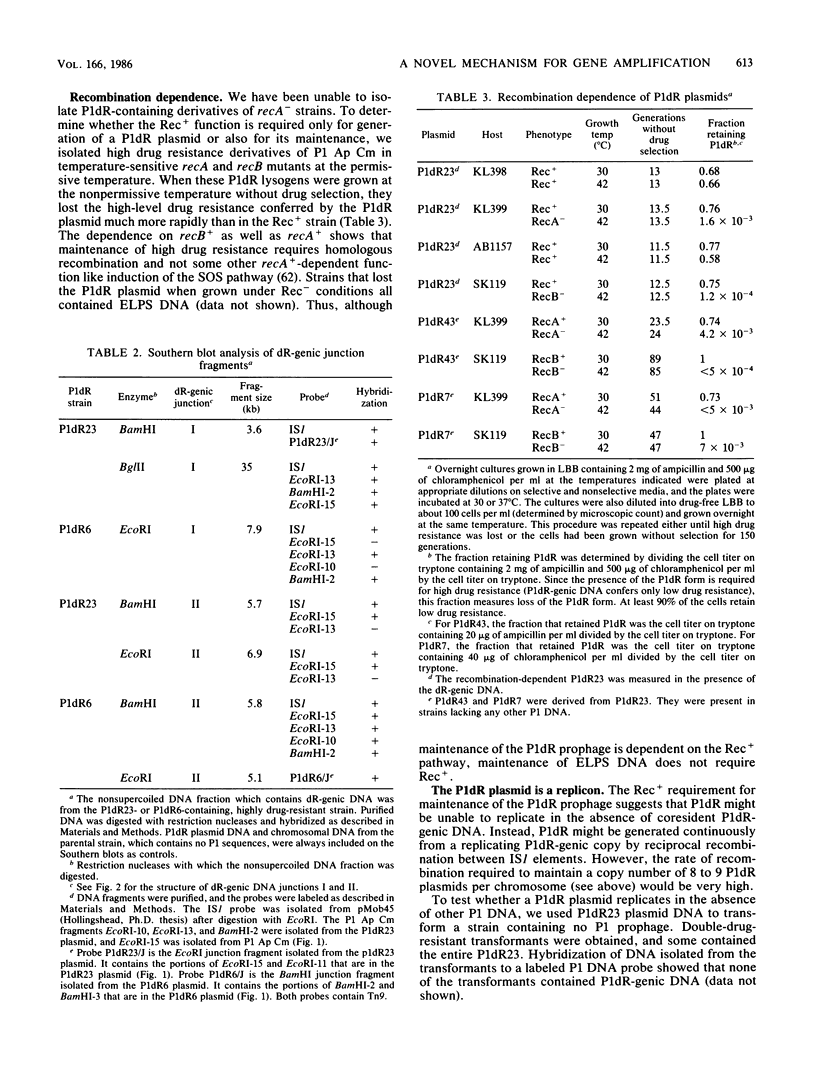



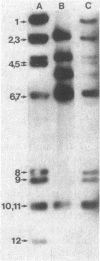
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abeles A. L., Snyder K. M., Chattoraj D. K. P1 plasmid replication: replicon structure. J Mol Biol. 1984 Mar 5;173(3):307–324. doi: 10.1016/0022-2836(84)90123-2. [DOI] [PubMed] [Google Scholar]
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Anderson R. P., Roth J. R. Tandem genetic duplications in phage and bacteria. Annu Rev Microbiol. 1977;31:473–505. doi: 10.1146/annurev.mi.31.100177.002353. [DOI] [PubMed] [Google Scholar]
- Austin S., Hart F., Abeles A., Sternberg N. Genetic and physical map of a P1 miniplasmid. J Bacteriol. 1982 Oct;152(1):63–71. doi: 10.1128/jb.152.1.63-71.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumstark B. R., Lowery K., Scott J. R. Location by DNA sequence analysis of cop mutations affecting the number of plasmid copies of prophage P1. Mol Gen Genet. 1984;194(3):513–516. doi: 10.1007/BF00425567. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capage M. A., Scott J. R. SOS induction by P1 Km miniplasmids. J Bacteriol. 1983 Aug;155(2):473–480. doi: 10.1128/jb.155.2.473-480.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M., Allet B., Gallay E., Boy de La Tour E., Caro L. Involvement of IS1 in the dissociation of the r-determinant and RTF components of the plasmid R100.1. Mol Gen Genet. 1977 Jun 24;153(3):289–295. doi: 10.1007/BF00431594. [DOI] [PubMed] [Google Scholar]
- Chandler M., Clerget M., Galas D. J. The transposition frequency of IS1-flanked transposons is a function of their size. J Mol Biol. 1982 Jan 15;154(2):229–243. doi: 10.1016/0022-2836(82)90062-6. [DOI] [PubMed] [Google Scholar]
- Chandler M., Silver L., Lane D., Caro L. Properties of an autonomous r-determinant from R100.1. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1223–1231. doi: 10.1101/sqb.1979.043.01.139. [DOI] [PubMed] [Google Scholar]
- Chandler M., Séchaud J., Caro L. A mutant of the plasmid R100.1 capable of producing autonomous circular forms of its resistance determinant. Plasmid. 1982 May;7(3):251–262. doi: 10.1016/0147-619x(82)90006-3. [DOI] [PubMed] [Google Scholar]
- Clerget M., Chandler M., Caro L. Isolation of an IS1 flanked kanamycin resistance transposon from R1drd19. Mol Gen Genet. 1980;180(1):123–127. doi: 10.1007/BF00267360. [DOI] [PubMed] [Google Scholar]
- Clerget M., Chandler M., Caro L. Isolation of the kanamycin resistance region (Tn2350) of plasmid R1drd-19 as an autonomous replicon. J Bacteriol. 1982 Aug;151(2):924–931. doi: 10.1128/jb.151.2.924-931.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerget M., Chandler M., Caro L. The structure of R1drd19: a revised physical map of the plasmid. Mol Gen Genet. 1981;181(2):183–191. doi: 10.1007/BF00268425. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G., Saedler H. Deletions and an inversion induced by a resident IS1 of the lactose transposon Tn951. Mol Gen Genet. 1980;178(2):367–374. doi: 10.1007/BF00270486. [DOI] [PubMed] [Google Scholar]
- Cowan J. A., Scott J. R. Incompatibility among group Y plasmids. Plasmid. 1981 Sep;6(2):202–221. doi: 10.1016/0147-619x(81)90067-6. [DOI] [PubMed] [Google Scholar]
- Edlund T., Normark S. Recombination between short DNA homologies causes tandem duplication. Nature. 1981 Jul 16;292(5820):269–271. doi: 10.1038/292269a0. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J., Schmeissner U., Hofer M., Miller J. H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978 Dec 25;126(4):847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Froehlich B. J., Tatti K., Scott J. R. Evidence for positive regulation of plasmid prophage P1 replication: integrative suppression by copy mutants. J Bacteriol. 1983 Oct;156(1):205–211. doi: 10.1128/jb.156.1.205-211.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Chandler M. Structure and stability of Tn9-mediated cointegrates. Evidence for two pathways of transposition. J Mol Biol. 1982 Jan 15;154(2):245–272. doi: 10.1016/0022-2836(82)90063-8. [DOI] [PubMed] [Google Scholar]
- Gelfand D. H., Shepard H. M., O'Farrell P. H., Polisky B. Isolation and characterization of ColE1-derived plasmid copy-number mutant. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5869–5873. doi: 10.1073/pnas.75.12.5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hashimoto H., Rownd R. H. Transition of the R factor NR1 and Proteus mirabilis: level of drug resistance of nontransitioned and transitioned cells. J Bacteriol. 1975 Jul;123(1):56–68. doi: 10.1128/jb.123.1.56-68.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. W., Grafstrom R. H., Harnish B. W., Hillman B. S. Tandem duplications resulting from recombination between ribosomal RNA genes in Escherichia coli. J Mol Biol. 1977 Nov 5;116(3):407–428. doi: 10.1016/0022-2836(77)90077-8. [DOI] [PubMed] [Google Scholar]
- Iida S., Arber W. Multiple physical differences in the genome structure of functionally related bacteriophages P1 and P7. Mol Gen Genet. 1979 Jun 20;173(3):249–261. doi: 10.1007/BF00268635. [DOI] [PubMed] [Google Scholar]
- Ikeda H., Tomizawa J. Prophage P1, and extrachromosomal replication unit. Cold Spring Harb Symp Quant Biol. 1968;33:791–798. doi: 10.1101/sqb.1968.033.01.091. [DOI] [PubMed] [Google Scholar]
- KONDO E., MITSUHASHI S. DRUG RESISTANCE OF ENTERIC BACTERIA. IV. ACTIVE TRANSDUCING BACTERIOPHAGE P1 CM PRODUCED BY THE COMBINATION OF R FACTOR WITH BACTERIOPHAGE P1. J Bacteriol. 1964 Nov;88:1266–1276. doi: 10.1128/jb.88.5.1266-1276.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Kushner S. R. In vivo studies of temperature-sensitive recB and recC mutants. J Bacteriol. 1974 Dec;120(3):1213–1218. doi: 10.1128/jb.120.3.1213-1218.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R. G., Low B., Godson G. N., Birge E. A. Isolation and characterization of an Escherichia coli K-12 mutant with a temperature-sensitive recA- phenotype. J Bacteriol. 1974 Oct;120(1):407–415. doi: 10.1128/jb.120.1.407-415.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luder A., Mosig G. Two alternative mechanisms for initiation of DNA replication forks in bacteriophage T4: priming by RNA polymerase and by recombination. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1101–1105. doi: 10.1073/pnas.79.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinkoth J., Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984 May 1;138(2):267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- Meyer J., Iida S. Amplification of chloramphenicol resistance transposons carried by phage P1Cm in Escherichia coli. Mol Gen Genet. 1979 Oct 3;176(2):209–219. doi: 10.1007/BF00273215. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Ohtsubo E. Nucleotide sequence of an insertion element, IS1. Proc Natl Acad Sci U S A. 1978 Feb;75(2):615–619. doi: 10.1073/pnas.75.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D., Rownd R. H. Transition of R factor NR1 in Proteus mirabilis: molecular structure and replication of NR1 deoxyribonucleic acid. J Bacteriol. 1975 Sep;123(3):1013–1034. doi: 10.1128/jb.123.3.1013-1034.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson B. C., Rownd R. H. Drug resistance gene amplification of plasmid NR1 derivatives with various amounts of resistance determinant DNA. J Bacteriol. 1985 Mar;161(3):1042–1048. doi: 10.1128/jb.161.3.1042-1048.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P., Chandler M., Caro L. Replication of prophage P1 during the cell cycle of Escherichia coli. Mol Gen Genet. 1977 Mar 28;152(1):71–76. doi: 10.1007/BF00264942. [DOI] [PubMed] [Google Scholar]
- Rownd R., Mickel S. Dissociation and reassociation of RTF and r-determinants of the R-factor NR1 in Proteus mirabilis. Nat New Biol. 1971 Nov 10;234(45):40–43. doi: 10.1038/newbio234040a0. [DOI] [PubMed] [Google Scholar]
- Scott J. R. A turbid plaque-forming mutant of phage P1 that cannot lysogenize Escherichia coli. Virology. 1974 Dec;62(2):344–349. doi: 10.1016/0042-6822(74)90397-3. [DOI] [PubMed] [Google Scholar]
- Scott J. R. A turbid plaque-forming mutant of phage P1 that cannot lysogenize Escherichia coli. Virology. 1974 Dec;62(2):344–349. doi: 10.1016/0042-6822(74)90397-3. [DOI] [PubMed] [Google Scholar]
- Scott J. R., Kropf M. M., Padolsky L., Goodspeed J. K., Davis R., Vapnek D. Mutants of plasmid prophage P1 and elevated copy number: isolation and characterization. J Bacteriol. 1982 Jun;150(3):1329–1339. doi: 10.1128/jb.150.3.1329-1339.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev N., Cohen G. Control of circularization of bacteriophage P1 DNA in Escherichia coli. Virology. 1981 Oct 30;114(2):333–342. doi: 10.1016/0042-6822(81)90215-4. [DOI] [PubMed] [Google Scholar]
- Shepard H. M., Gelfand D. H., Polisky B. Analysis of a recessive plasmid copy number mutant: evidence for negative control of Col E1 replication. Cell. 1979 Oct;18(2):267–275. doi: 10.1016/0092-8674(79)90046-1. [DOI] [PubMed] [Google Scholar]
- Silver L., Chandler M., Lane H. E., Caro L. Production of extrachromosomal r-determinant circles from integrated R100.1: involvement of the E. coli recombination system. Mol Gen Genet. 1980;179(3):565–571. doi: 10.1007/BF00271746. [DOI] [PubMed] [Google Scholar]
- Smith G. E., Summers M. D. The bidirectional transfer of DNA and RNA to nitrocellulose or diazobenzyloxymethyl-paper. Anal Biochem. 1980 Nov 15;109(1):123–129. doi: 10.1016/0003-2697(80)90019-6. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sternberg N., Austin S. Isolation and characterization of P1 minireplicons, lambda-P1:5R and lambda-P1:5L. J Bacteriol. 1983 Feb;153(2):800–812. doi: 10.1128/jb.153.2.800-812.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N., Austin S. The maintenance of the P1 plasmid prophage. Plasmid. 1981 Jan;5(1):20–31. doi: 10.1016/0147-619x(81)90075-5. [DOI] [PubMed] [Google Scholar]
- Sternberg N. Demonstration and analysis of P1 site-specific recombination using lambda-P1 hybrid phages constructed in vitro. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1143–1146. doi: 10.1101/sqb.1979.043.01.128. [DOI] [PubMed] [Google Scholar]
- Taylor D. P., Greenberg J., Rownd R. H. Generation of miniplasmids from copy number mutants of the R plasmid NR1. J Bacteriol. 1977 Dec;132(3):986–995. doi: 10.1128/jb.132.3.986-995.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin B. E., Nordström K. R plasmid gene dosage effects in Escherichia coli K-12: copy mutants of the R plasmic R1drd-19. Plasmid. 1977 Nov;1(1):1–7. doi: 10.1016/0147-619x(77)90003-8. [DOI] [PubMed] [Google Scholar]
- West B. W., Scott J. R. Superinfection immunity and prophage repression in phage P1 and P7. III. Induction by virulent mutants. Virology. 1977 May 1;78(1):267–276. doi: 10.1016/0042-6822(77)90098-8. [DOI] [PubMed] [Google Scholar]
- Witkin E. M. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol Rev. 1976 Dec;40(4):869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun T., Vapnek D. Electron microscopic analysis of bacteriophages P1, P1Cm, and P7. Determination of genome sizes, sequence homology, and location of antibiotic-resistance determinants. Virology. 1977 Mar;77(1):376–385. doi: 10.1016/0042-6822(77)90434-2. [DOI] [PubMed] [Google Scholar]
- Zabrovitz S., Segev N., Cohen G. Growth of bacteriophage P1 in recombination-deficient hosts of Escherichia coli. Virology. 1977 Jul 15;80(2):233–248. doi: 10.1016/s0042-6822(77)80001-9. [DOI] [PubMed] [Google Scholar]