Abstract
The sfrB gene of Escherichia coli K-12 and the rfaH gene of Salmonella typhimurium LT2 are homologous, controlling expression of the tra operon of F and the rfa genes for lipopolysaccharide synthesis. We have determined a restriction map of the 19-kilobase ColE1 plasmid pLC14-28 which carries the sfrB gene of E. coli. After partial Sau3A digestion of pLC14-28, we cloned a 2.5-kilobase DNA fragment into the BamHI site of pBR322 to form pKZ17. pKZ17 complemented mutants of the sfrB gene of E. coli and the rfaH gene of S. typhimurium for defects of both the F tra operon and the rfa genes. pKZ17 in minicells determines an 18-kilodalton protein not determined by pBR322. A Tn5 insertion into the sfrB gene causes loss of complementing activity and loss of the 18-kilodalton protein in minicells, indicating that this protein is the sfrB gene product. These data indicate that the sfrB gene product is a regulatory element, since the single gene product elicits the expression of genes for many products for F expression and lipopolysaccharide synthesis.
Full text
PDF

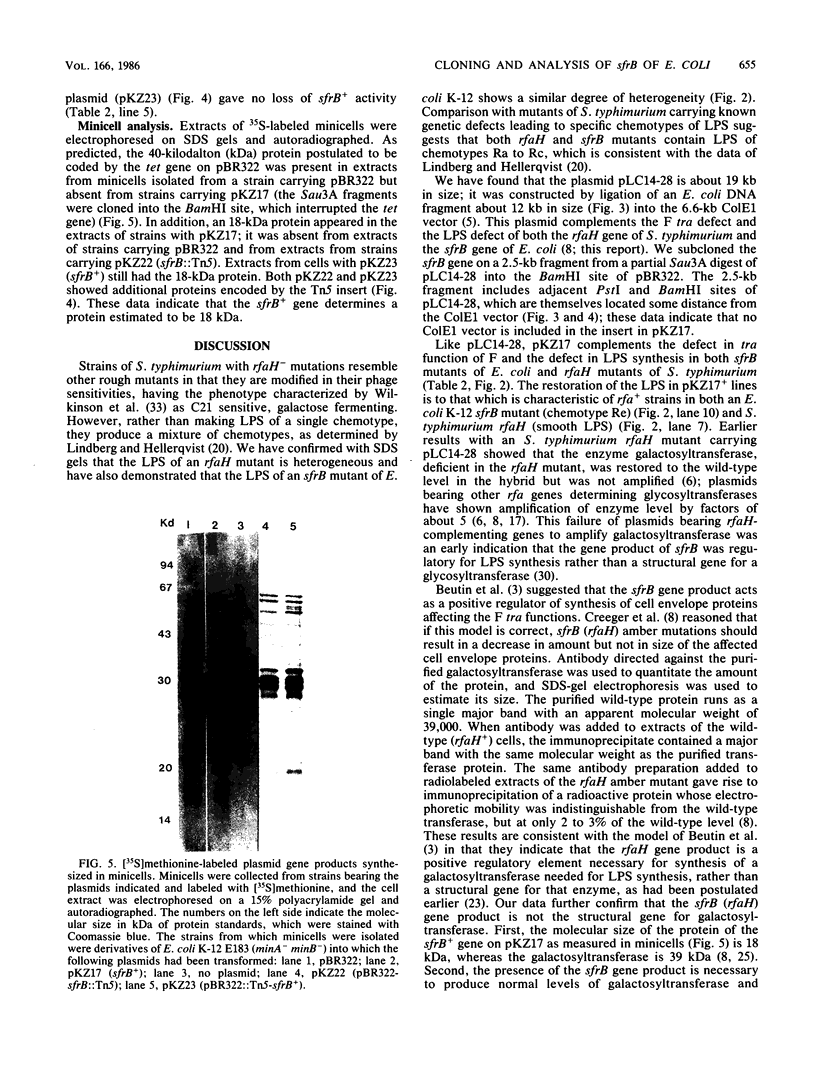


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson E. S., Smith H. R. Fertility inhibition in strains of Salmonella typhimurium. Mol Gen Genet. 1972;118(1):79–84. [PubMed] [Google Scholar]
- Beutin L., Achtman M. Two Escherichia coli chromosomal cistrons, sfrA and sfrB, which are needed for expression of F factor tra functions. J Bacteriol. 1979 Sep;139(3):730–737. doi: 10.1128/jb.139.3.730-737.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutin L., Manning P. A., Achtman M., Willetts N. sfrA and sfrB products of Escherichia coli K-12 are transcriptional control factors. J Bacteriol. 1981 Feb;145(2):840–844. doi: 10.1128/jb.145.2.840-844.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco C., Mata-Gilsinger M., Ritzenthaler P. Construction of hybrid plasmids containing the Escherichia coli uxaB gene: analysis of its regulation and direction of transcription. J Bacteriol. 1983 Feb;153(2):747–755. doi: 10.1128/jb.153.2.747-755.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976 Sep;9(1):91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Creeger E. S., Chen J. F., Rothfield L. I. Cloning of genes for bacterial glycosyltransferases. II. Selection of a hybrid plasmid carrying the rfah gene. J Biol Chem. 1979 Feb 10;254(3):811–815. [PubMed] [Google Scholar]
- Creeger E. S., Rothfield L. I. Cloning of genes for bacterial glycosyltransferases. I. Selection of hybrid plasmids carrying genes for two glucosyltransferases. J Biol Chem. 1979 Feb 10;254(3):804–810. [PubMed] [Google Scholar]
- Creeger E. S., Schulte T., Rothfield L. I. Regulation of membrane glycosyltransferases by the sfrB and rfaH genes of Escherichia coli and Salmonella typhimurium. J Biol Chem. 1984 Mar 10;259(5):3064–3069. [PubMed] [Google Scholar]
- Finnegan D. J., Willetts N. S. Two classes of Flac mutants insensitive to transfer inhibition by an F-like R factor. Mol Gen Genet. 1971;111(3):256–264. doi: 10.1007/BF00433110. [DOI] [PubMed] [Google Scholar]
- Foulds J., Barrett C. Characterization of Escherichia coli mutants tolerant to bacteriocin JF246: two new classes of tolerant mutants. J Bacteriol. 1973 Nov;116(2):885–892. doi: 10.1128/jb.116.2.885-892.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin N. C., Bennett G. N. The N protein of bacteriophage lambda, defined by its DNA sequence, is highly basic. Gene. 1979 Dec;8(1):107–119. doi: 10.1016/0378-1119(79)90011-8. [DOI] [PubMed] [Google Scholar]
- Gaffney D., Skurray R., Willetts N. Regulation of the F conjugation genes studied by hybridization and tra-lacZ fusion. J Mol Biol. 1983 Jul 25;168(1):103–122. doi: 10.1016/s0022-2836(83)80325-8. [DOI] [PubMed] [Google Scholar]
- Greenblatt J., Li J. Properties of the N gene transcription antitermination protein of bacteriophage lambda. J Biol Chem. 1982 Jan 10;257(1):362–365. [PubMed] [Google Scholar]
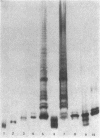
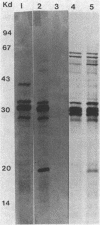
- Hitchcock P. J., Brown T. M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983 Apr;154(1):269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R. T., Koeltzow D. E., Stocker B. A. Genetic transfer of Salmonella typhimurium and Escherichia coli lipopolysaccharide antigens to Escherichia coli K-12. J Bacteriol. 1972 Sep;111(3):758–770. doi: 10.1128/jb.111.3.758-770.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam S. K., Rehemtulla A., Sanderson K. E. Cloning of rfaG, B, I, and J genes for glycosyltransferase enzymes for synthesis of the lipopolysaccharide core of Salmonella typhimurium. J Bacteriol. 1985 Jan;161(1):277–284. doi: 10.1128/jb.161.1.277-284.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T. T., Stocker B. A. Mapping of rfa Genes in Salmonella typhimurium by ES18 and P22 Transduction and by Conjugation. J Bacteriol. 1972 Oct;112(1):48–57. doi: 10.1128/jb.112.1.48-57.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lindberg A. A., Hellerqvist C. G. Rough mutants of Salmonella typhimurium: immunochemical and structural analysis of lipopolysaccharides from rfaH mutants. J Gen Microbiol. 1980 Jan;116(1):25–32. doi: 10.1099/00221287-116-1-25. [DOI] [PubMed] [Google Scholar]
- Osborn M. J. Biochemical characterization of mutants of Salmonella typhimurium lacking glucosyl or galactosyl lipopolysaccharide transferases. Nature. 1968 Mar 9;217(5132):957–960. doi: 10.1038/217957a0. [DOI] [PubMed] [Google Scholar]
- Roantree R. J., Kuo T. T., MacPhee D. G. The effect of defined lipopolysaccharide core defects upon antibiotic resistances of Salmonella typhimurium. J Gen Microbiol. 1977 Dec;103(2):223–234. doi: 10.1099/00221287-103-2-223. [DOI] [PubMed] [Google Scholar]
- Romeo D., Hinckley A., Rothfield L. Reconstitution of a functional membrane enzyme system in a monomolecular film. II. Formation of a functional ternary film of lipopolysaccharide, phospholipid and transferase enzyme. J Mol Biol. 1970 Nov 14;53(3):491–501. doi: 10.1016/0022-2836(70)90079-3. [DOI] [PubMed] [Google Scholar]
- Sanderson K. E., Kadam S. K., MacLachlan P. R. Derepression of F factor function in Salmonella typhimurium. Can J Microbiol. 1983 Sep;29(9):1205–1212. doi: 10.1139/m83-184. [DOI] [PubMed] [Google Scholar]
- Sanderson K. E., Roth J. R. Linkage map of Salmonella typhimurium, Edition VI. Microbiol Rev. 1983 Sep;47(3):410–453. doi: 10.1128/mr.47.3.410-453.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K. E., Saeed Y. A. P22-mediated transduction analysis of the rough A (rfa) region of the chromosome of Salmonella typhimurium. J Bacteriol. 1972 Oct;112(1):58–63. doi: 10.1128/jb.112.1.58-63.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandulache R., Prehm P., Kamp D. Cell wall receptor for bacteriophage Mu G(+). J Bacteriol. 1984 Oct;160(1):299–303. doi: 10.1128/jb.160.1.299-303.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker B. A., Males B. M., Takano W. Salmonella typhimurium mutants of RfaH-phenotype: genetics and antibiotic sensitivities. J Gen Microbiol. 1980 Jan;116(1):17–24. doi: 10.1099/00221287-116-1-17. [DOI] [PubMed] [Google Scholar]
- Wilkinson R. G., Gemski P., Jr, Stocker B. A. Non-smooth mutants of Salmonella typhimurium: differentiation by phage sensitivity and genetic mapping. J Gen Microbiol. 1972 May;70(3):527–554. doi: 10.1099/00221287-70-3-527. [DOI] [PubMed] [Google Scholar]