Abstract
Intracellular trafficking of major histocompatibility complex (MHC) class II molecules is characterized by passage through specialized endocytic compartment(s) where antigenic peptides replace invariant chain fragments in the presence of the DM protein. These changes are accompanied by structural transitions of the MHC molecules that can be visualized by formation of compact SDS-resistant dimers, by changes in binding of mAbs, and by changes in T cell responses. We have observed that a mAb (25-9-17) that is capable of staining I-Ab on the surface of normal B cells failed to interact with I-Ab complexes with a peptide derived from the Eα chain of the I-E molecule but bound a similar covalent complex of I-Ab with the class II binding fragment (class II-associated invariant chain peptides) of the invariant chain. Moreover, 25-9-17 blocked activation of several I-Ab-reactive T cell hybridomas but failed to block others, suggesting that numerous I-Ab-peptide complexes acquire the 25-9-17+ or 25-9-17− conformation. Alloreactive T cells were also able to discriminate peptide-dependent variants of MHC class II molecules. Thus, peptides impose subtle structural transitions upon MHC class II molecules that affect T cell recognition and may thus be critical for T cell selection and autiommunity.
It has been appreciated that major histocompatibility complex (MHC) class II molecules undergo conformational changes during their transport to the cell surface. These changes were detected as changes in mAb epitopes (1, 2, 3) or the ability to acquire stability in SDS (4). Another important factor in the structural transitions of MHC class II molecules appears to be the hydrogen ion concentration. A weakly acidic environment causes a loss of SDS stability and enhances the binding of 1-anilino-naphthalene-8-sulfonic acid, which is a marker for exposed hydrophobic sites (5, 6). Acidic pH enhances peptide binding (7–9) and is optimal for class II-associated invariant chain peptides (CLIP)/peptide exchange catalyzed by HLA-DM molecules (10, 11), suggesting that protonation of certain residues in the MHC class II molecule may cause transient conformational shifts that allow optimal peptide binding and/or exchange.
Whether the mature MHC class II molecules expressed on the surface of antigen-presenting cells exist in different conformations relevant to T cell recognition remains unclear. It is well appreciated that peptides are able to change the conformation of MHC class I molecules. These changes were detected as gain/loss of binding by anti-class I antibodies (12–15) and by analysis of MHC class I molecules crystallized with single peptides (16, 17). We sought to determine whether peptide-dependent changes occur in MHC class II that could be detected by mAbs. While analyzing anti-MHC class II mAb staining of cells expressing MHC class II complexes with single peptides, we found that mAb 25-9-17, which reacts with I-Ab, fails to bind a complex between I-Ab and Eα peptide. This observation prompted us to seek explanations for this phenomenon. Indeed, we found that MHC class II molecules available for T cell recognition have subtle conformational differences dependent on the particular peptide they bind and that these alterations can also be detected by T cells.
MATERIALS AND METHODS
Mice.
C57BL6/J (B6), B10.A-H2i5H2-Tlaa(5R)/SgSnEg (5R), and B6.C-H2bm12/KhEg (bm12) were obtained from The Jackson Laboratory. Invariant chain (Ii) knock-out animals were a gift from R.Flavell and I. Shachar (Yale University). Transgenic mice expressing covalently bound Eα peptide (Ab–pEα)were a gift from P. Marrack (National Jewish Center, Denver, CO). H2-M-deficient mice were provided by Luc Van Kaer (Vanderbilt University, Nashville, TN).
Cells and Tissue Culture.
All cell cultures were performed in complete medium [Click’s EHAA medium (Irvine Scientific) with 5% fetal calf serum (Intergen, Purchase, NY), 5 × 10−5 M 2-mercaptoethanol (Bio-Rad), 2 mM l-glutamine, 50 μg/ml gentamicin (GIBCO/BRL)].
The bm12-anti-Ii KO T cell lines were produced by stimulation in vitro of purified CD4 T cells with 2,000-R irradiated Ii KO splenocytes (1 R = 0.258 mC/kg) as described (18). Cell lines were sustained by restimulation with Ii KO splenocytes every 10–14 days. To purify CD4+ T cells bm12 lymph node cells were treated with a mixture of anti-MHC class II and anti-CD8 mAbs followed by a mixture of magnetic beads conjugated to anti-mouse IgG, anti-mouse IgM, and anti-rat IgG (PerSeptive Biosystems, Cambridge, MA). T cell hybridomas were produced by fusion of T cell lines (day 5–7 after activation) with T cell lymphoma BW5147 using polyethylene glycol (Mr =1500; Boehringer Mannheim) (19).
D10 T cell clone (20) and bm12-anti Ii KO T cell line proliferation stimulated with allo-MHC was tested in duplicate or triplicate cultures in a total volume of 200 μl in flat-bottom 96-well plates (Becton-Dickinson). Responder cells were used at 2 × 104 cells per well and 105 irradiated (2,000 rads; 1 rad = 0.01 Gy) spleen cells or 1–3 × 103 L cells treated (100 μg/ml for 1 hr at 37°C) with mitomycin C (Sigma) were used as stimulators. Proliferation was assessed by [3H]thymidine incorporation after 48 hr of incubation at 37°C. Proliferation of the bm12-anti Ii KO T cell line against transfected L cells was estimated in the presence of interleukin 2 (IL-2, 5 units/ml) and the background proliferation in the presence of untransfected L cells and IL-2 was subtracted from the [3H]thymidine incorporation.
IL-2 production by Ab-pEα-specific hybridoma 1H3.1 (21) and others has been tested by proliferation of the IL-2-dependent CTLL cell line measured either by [3H]thymidine incorporation or by colorimetric assay using Alamar blue (AccuMed, Westlake, OH) according to manufacturer’s instructions. H-2M knock-out splenocytes were preincubated with peptides for 2–3 hr before addition of blocking antibodies.
mAbs.
Hybridomas producing anti-MHC class II mAb 25–9-17 (22) and M/5114 (23), as well as anti-CD8 (53–6.72) (24), were purchased from the American Type Culture Collection; Y3JP (25), YAe (26) and CerCLIP.1 (27) were produced at Yale University. Antibodies were used as supernatants or as purified proteins. Antibody purification was performed by protein-G affinity chromatography by the Immunobiology Hybridoma Facility.
Flow Cytometry (FACS) Analysis.
Cells for FACS analysis were stained in microtiter U-bottom plates in 50 μl of supernatants for 30 min on ice, followed by secondary staining with goat anti-mouse Ig-fluorescein isothiocyanate-conjugate(Fc-specific; Sigma) for 30 min on ice in PBS containing 2%fetal calf serum and 0.1%NaN3. Splenic B cells were counterstained with anti-B220 mAb conjugated to phycoerythrin (PharMingen). Cells were analyzed on FACScan flow cytometer (Becton-Dickinson) using cellquest software.
DNA Constructs and Transfection.
Construct encoding Abβ with covalently bound peptide Eα52–68 (Ab-pEα) was a gift from P. Marrack. The Ab–murine CLIP (mCLIP) construct was made as follows: poly-Gly linker region was shortened by 3 amino acids by introduction of a new BamHI site, cutting with BamHI, and religation. Oligonucleotide containing CLIP 83–103 sequence, a part of the linker and in-frame NheI and BamHI sites was used for cloning into the Ab-pEα construct from which the NheI–BamHI fragment was removed. L cells were stably transfected with MHC class II α and β chain cDNA and a neo-encoding cDNA by the calcium phosphate precipitation method (28). Transient transfections were performed with the DEAE-dextran method exactly as described (29). Ab-pEα-expressing L cells were supertransfected with a p31 Ii cDNA (a gift from Jim Miller, University of Chicago) along with a plasmid containing the thymidine kinase gene. Negative selection of nontransfected cells was performed by cultivation in G418 and hypoxanthine/aminopterin/thymidine (HAT) medium (both from GIBCO/BRL).
Western Blotting and Immunoprecipitations.
For Western blotting, splenocytes were lysed as described above and lysates were precleared with anti-mouse Ig-agarose (Sigma) to remove endogenous Igs. Unboiled proteins were separated by SDS/PAGE on 8% gels and transferred onto nitrocellulose membrane. MHC class II was stained with mAbs YAe (26), Y3JP(25), and 25-91-7(23), followed by horseradish peroxidase-coupled anti-mouse Ig antibodies (Bio-Rad) and ECL Western blotting detection reagent (Amersham) followed by exposure to x-ray film.
The SDS-stability assay was performed as described (10). In brief, mixtures of 2–4 × 104 cpm of affinity-purified Ab–human CLIP (hCLIP) complexes (metabolically labeled with [35S]methionine) isolated from T2.I-Ab cell line with the help of CerCLIP.1 mAb, DM-Sepharose beads, and 100 μM peptide was incubated at pH 5.0 for 2 hr at 37°C in the presence of 0.1% C12E9 (Sigma). The mixtures were then neutralized by addition of 1 M Tris and precipitated with mAb and protein-G Sepharose. Samples were eluted with nonreducing Laemmli sample buffer and separated by SDS/PAGE on 10% gels without boiling. An Ak-binding peptide HEL 46–61 (NTDGSTDYGILQINSRY) and Ab-binding Eα 52–68 peptide(ASFEAQGALANIAVDKA) (21, 30) were synthesized by the Keck Foundation Biotechnology Resource Laboratory (Yale University). Peptides indicated in Table 1 were synthesized on a Synergy 432 automated peptide synthesizer (Applied Biosystems) using fluorenylmethoxycarbonyl chemistry. The purity of peptides used was greater than 90%.
Table 1.
mAB 25-9-17 shows selective recognition of some Ab–peptide complexes
| % inhibition of T cells response by mAb
|
|||
|---|---|---|---|
| Peptide [source] | Y3JP | 25-9-17 | YAe |
| Endogenous peptides* | |||
| Unknown +Ab (D10 allo) | 99 | 99.5 | 0 |
| SAKPVSQMRMATPLLMRPMSM (CLIP) + linker + Ab | 99 | 98 | 0 |
| Unknown +Ab (D10 allo), Ii KO | 95 | 73.5 | 0 |
| Exogenously added peptides† | |||
| RNIYWTDSVPGSVSVA [LDL receptor, residues 486–501] | 100 | 100 | 0 |
| TQFHPPHIEIQ [β2-microglobulin, residues 48–58] | 100 | 100 | 0 |
| ITNQYNSPTGLYSSKN [Clp36, residues 138–154] | 100 | 49 | 0 |
| VPIYEGYALPHAILR [β-actin, residues 48–56] | 100 | 38 | 0 |
| NLCNIPASALLSSDIP [hen-egg lysozyme, residues 74–88] | 100 | 43 | 0 |
| APMYYRGAQAAIVVYD [Rab5, residues 86–101] | 100 | 52 | 0 |
| ASFEAQGALANIAVDKA [Eα52–68]‡ | 97.4 | 0 | 97 |
| TKNLDYVATSIHEAVT [aspartate aminotransferase, residues 396–410] | 98 | −3.5 | 0 |
| GENPIYKSAVTTVVNPK [integrin β1, residues 778–794] | 100 | 8.5 | 0 |
| GQNAWFLPAQADIVATK [CD98, residues 207–223] | 100 | −9 | 0 |
| DWTVDHPQTLFAWEG [CD22, residues 25–39] | 95 | 16 | 0 |
Proliferation of D10 T cells in response to endogenous peptides was blocked by addition of purified mAbs (5 μg/ml). Percent of inhibition is shown relative to addition of equal concentration of YAe as a negative control. Mean of two separate experiments.
Inhibition of IL-2 secretion by T cell hybridomas, measured by proliferation of IL-2-dependent cells, was estimated. T cell hybridomas were generated against synthetic analogues of peptides presented naturally by Ab (refs. 13 and 24 and A.Y.R., unpublished results). Data are from representative experiments. Inhibition was calculated relative to equal concentrations of the anti-class I mAb Y-3. Both peptides and antibodies were titrated. The data for 25-9-17 represent maximal achievable inhibition. This was measured at a peptide concentration when the response is clearly detectable and is below saturation and at a maximal concentration of mAb showing inhibition. The data for Y3JP and YAe are shown for the same peptide and mAb concentrations as for 25-9-17 for every T cell.
1.H3.1 T cell hybridoma (13) was used to test anti-Ab–pEα response.
RESULTS
Mature MHC-Class II I-Ab Molecules Can Accommodate More Than One Conformational State Depending on the Bound Peptide.
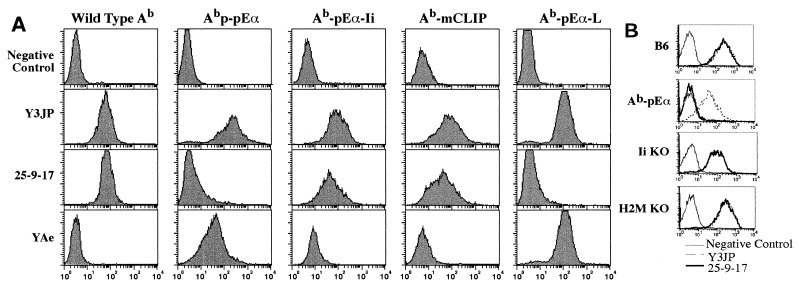
mAbs have been used as a powerful tool to assess conformational changes in MHC class II molecules during their biosynthesis and folding (1–3). To determine whether the differences in conformation of MHC class II molecules may be attributed to the nature of their associated bound peptides, we used a number of anti-I-Ab mAb to stain the surface of MHC class II expressing cells. The mouse fibroblast-like L cell line Ltk−, which is negative for Ii, was used in our experiments because Ii has been shown to influence MHC class II antibody epitopes (2) and to remove the covalently bound peptides from MHC class II molecules (31). L cells stably were transfected with either wild-type (WT) Ab (AαbAβb) or Aαb plus modified Aβb. These modifications included Aβb encoding a covalently bound Eα peptide (pEα52–68) (31) (courtesy of P. Marrack), mouse CLIP (mCLIP; a fragment of mouse Ii residues 83–103), and Eα peptide with the poly-Gly linker of the same length as in mCLIP construct (Ab-pEα-L). Surface expression of MHC class II encoded by the described genes was analyzed by FACS (Fig. 1) using various mAb with specificity for I-Ab: Y3JP (25), 25-9-17 (22), and YAe (26), which only recognizes the Ab–pEα complex (21). It is clear that mAb 25-9-17 failed to bind Ab–pEα but did bind WT Ab or Ab-mCLIP molecules, suggesting that the epitope recognized by this antibody may be sensitive to conformational changes imposed by one set of peptides but not by other sets (Fig. 1A). Similar results were obtained with splenic B cells from I-Ab-pEα transgenic animals (Fig. 1B). The analysis of B cells (electronically gated after double-staining with anti-class II and anti-B220) has shown that the entire B cell population was either 25-9-17-positive (in B6, Ii KO, and H2M KO) or 25-9-17-negative (in Ab-pEα transgenic mice). The data suggest that 25-9-17+ and 25-9-17− complexes must be expressed by the same cell.
Figure 1.
Conformational variants of the MHC class II imposed by different peptides are detected on the cell surface by mAb. (A) Stable bulk populations of indicated L cell transfectants were stained with anti-class II mAb followed by anti-Ig-fluorescein isothiocyanate-conjugated antibodies and analyzed by FACS. Ab-pEα and Ab-mCLIP are cotransfectants of Abα and Abβ covalently linked to pEα52–68 or mouse CLIP83–103, respectively. Ab-pEα-Ii is Ab-pEα line supertransfected with Ii cDNA. In the Ab-Eα-L line the length of the poly-Gly linker matches the linker in Ab-mCLIP. (B) FACS analysis of splenocytes from wild type B6 and indicated mutant mice. Cells were stained with anti-class II mAb, followed by secondary fluorescein isothiocyanate-conjugated anti-mouse Ig (Fc-specific) antibodies and rat anti-B220-phycoerythrin conjugate. MHC class II profiles are shown for electronically gated B220-positive B cells.
The lack of reactivity of 25-9-17 with Ab–pEα is could be due to the interference by the poly-Gly linker. To disprove the latter possibility, we made another transfectant, Ab-pEαL, that has the same linker as Ab-mCLIP; this transfectant showed the same lack of staining as the original Ab-pEα (Fig. 1A). Thus, the MHC conformation that doesn’t bind mAb 25-9-17 is determined by peptide binding rather than by the covalent linkage or by linker interference with mAb binding.
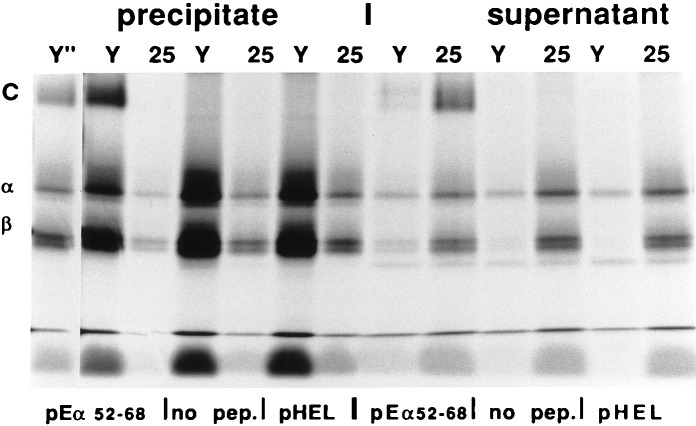
Furthermore, to test whether the observed phenomenon could be reproduced with noncovalent MHC-peptide complexes, we performed the following experiments. I-Ab molecules from the HLA-DM-deficient mutant T2 cell line have been shown to contain fragments of hCLIP that do not allow MHC class II to form SDS-stable compact dimers (1, 32). Metabolically labeled Ab-hCLIP complexes from the T2 cell line transfected with AαbAβb were affinity-purified. In the presence of purified DM molecules, hCLIP is exchanged for another peptide (10, 11), and compact dimer formation can be detected by examining these molecules with SDS/PAGE. Ab-hCLIP complexes were loaded with pEα 52–68 or control peptide (HEL46–61 fragment that does not efficiently bind Ab) and precipitated with mAb Y3JP or 25-9-17. It is evident (Fig. 2) that the SDS-stable dimers could be precipitated with Y3JP but not with 25-9-17. By contrast, compact dimers could be found in the unbound material phase of the immunoprecipitation reaction with 25-9-17 mAb, although they were largely depleted by mAb Y3JP. This finding demonstrates that the lack of binding of mAb 25-9-17 is not due to interference by the covalent linker, as in this case there was no linker, and also directly shows that interaction of mAb 25-9-17 with I-Ab depends upon the particular peptide bound by the peptide-binding domain.
Figure 2.
Failure of mAb 25-9-17 to interact with Ab–pEα is not due to interference by poly-Gly linker: this mAb also fails to precipitate Ab molecules loaded with pEα in vitro. Ab–hCLIP molecules metabolically labeled with [35S]methionine and affinity-purified from T2 cell line were reloaded with peptides in vitro in the presence of HLA-DM and precipitated with mAb Y3JP (lanes Y) and 25-9-17 (lanes 25) (PRECIPITATE). Part of the supernatant from the precipitation reaction was also electrophoresed on the gel (SUPERNATANT), which shows material nonabsorbed by a mAb. SDS-stable C dimers were only detected in the presence of Eα 52–68 peptide but not if no peptide or HEL 46–61 (does not bind Ab) were used. Because Y3JP appears to be a better binder than 25-9-17, a shorter exposure of the Y3JP precipitation of Ab-pEα is shown (Y" lane at far left).
To demonstrate that other MHC–peptide complexes can be discriminated on the basis of 25-9-17 reactivity, we used a panel of T cell hybridomas specific to recently identified natural Ab peptides (ref. 33 and A.Y.R., unpublished results). T cell reactivity against synthetic analogs of the natural peptides loaded onto H-2M knock-out splenocytes was tested in the presence of mAb (Table 1, exogenously added peptides). It is quite clear that mAb 25-9-17 binds (blocks) multiple Ab-peptide complexes and also fails to bind to (block) another set. mAb YAe is known to be specific for the Ab-pEα complex and binds no others, whereas mAb Y3JP did not discriminate among these complexes. Thus, multiple MHC class II–peptide complexes may have 25-9-17-negative conformation.
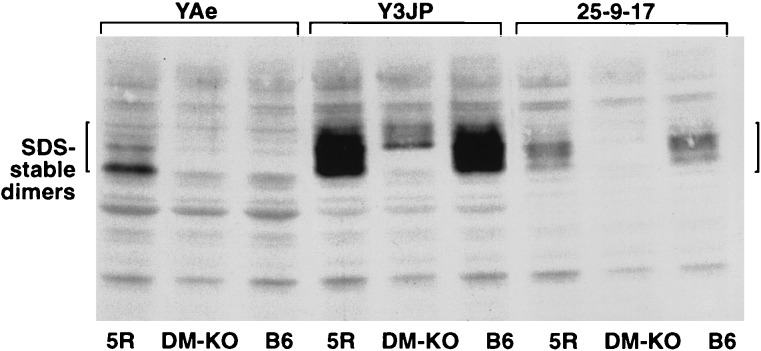
Most of the MHC class II molecules on the surface of H-2M and Ii sufficient splenocytes are in an SDS-stable compact form (4). To determine whether 25-9-17 interacts with compact dimers, lysates of I-Ab+ cells were analyzed by Western blotting. Nonboiled lysates of splenocytes from B6 (I-E negative), B10A.5R (Ab+, E+), and H2-M-knock-out (Ab+) were probed with Y3JP, 25-9-17, and YAe antibodies (Fig. 3). None of these mAb (Y3JP, 25-9-17, or YAe) interacted with free MHC class II chains (data not shown). Y3JP mAb interacted with SDS-stable dimers from all these sources, staining a relatively wide band. In H-2M-negative spleen, the floppy CLIP-bound Ab runs at the very top of the broad band stained by Y3JP in B6 I-Ab (Fig. 3). mAb YAe exclusively stained the C dimer in Eα-positive strain B10.A(5R). YAe+ band corresponds to the lower part of the wide Y3JP+ SDS-stable band. mAb 25-9-17 also stained some C dimers from Ab -positive spleens. Interaction of this mAb with SDS-stable dimers from H2-M-knock-out splenocytes was extremely weak and could only be detected upon longer exposure (data not shown). Staining of B6 and H-2M-knock-out spleens with serial dilutions of 25-9-17 suggested that the antibody has lower affinity for Ab in H-2M knock-outs than in WT animals (data not shown). Under the conditions of Western blotting (saturating amount of mAb), mAb 25-9-17 has the highest affinity to compact dimers positioned in the middle of the wide band stained with Y3JP.
Figure 3.
mabs Y3JP, 25-9-17 and YAe interact with mature MHC class II molecules (C dimers). Western blot analysis of SDS-stable MHC class II dimers from splenocytes from 5R, B6-H-2MA− (DM KO), and B6 mice.
Thus, SDS-stable dimers are heterogeneous and their relative mobility ranges from the most “tight” (corresponding to Ab-pEα complex detected by YAe) to the floppiest form detected by Y3JP in H2-M-knock-out class II. These differences in mobility of the SDS-stable complexes are too significant to be explained by variation in the length of bound peptides and are not related to differential glycosylation (data not shown). Rather, they reflect the slight differences in folding (conformational variants) of the MHC–peptide complexes that depend on the structure of the bound peptides. mAb 25-9-17 fails to interact with some of these complexes, and this failure is not restricted to a single Ab-pEα complex.
T Cells Discriminate 25-9-17+ and 25-9-17− MHC Class II Conformations.
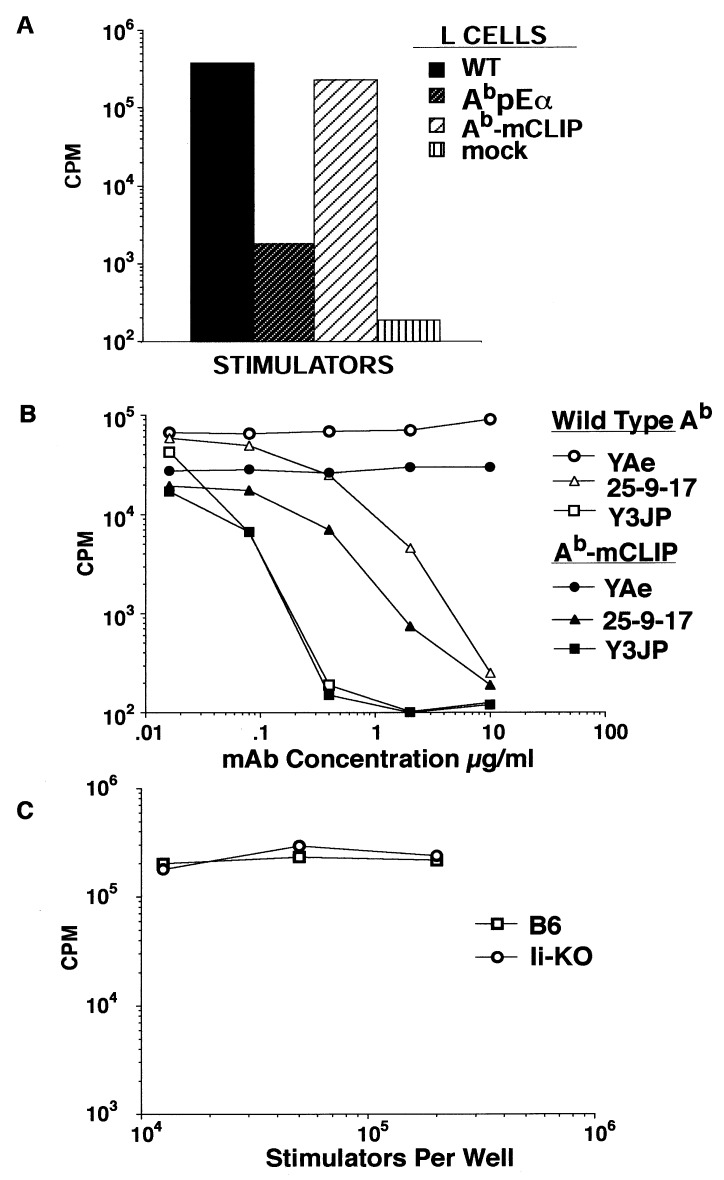
To analyze the ability of T cells to discriminate different conformational states of the MHC class II molecule, we performed a series of T cell activation assays. First, we investigated the capacity of L cells transfected with WT Ab, Ab-pEα, or Ab-mCLIP to stimulate the proliferative response of the D10 T cell clone, which is known to exhibit strong alloreactivity to Ab. As shown in Fig. 4A, D10 cells proliferate actively when stimulated with WT Ab or Ab-mCLIP but not by Ab-pEα. Reaction of D10 cells to both WT and Ab-mCLIP was blocked by anti-Ab mAbs including 25-9-17 (Fig. 4B). Thus, alloreactive D10 T cells recognize MHC class II molecules in the 25-9-17+ conformation. Interestingly, D10 cells react well to the WT Ab expressed by Ii-negative L cells and to Ab-mCLIP, suggesting that diverse peptides may be responsible for promotion of the 25-9-17+ conformation and D10 stimulation. D10 also responded well to splenocytes isolated from Ii-negative (knock-out) animals (Fig. 4C), and this reaction could be also blocked with mAb 25-9-17 (Table 1). Thus, T cell receptor recognition of the MHC class II complexes with diverse peptides is also sensitive to the conformational state of these complexes.
Figure 4.
T cells recognize variations in the conformations of the MHC class II-peptide complexes. (A) D10 T cells that recognize I-Ab with various undefined peptides proliferate in response to invariant chain negative L cells transfected with WT Ab and Ab-mCLIP but not Ab-pEα cDNA. (B) D10 reactivity to WT or Ab-mCLIP expressed by Ii-negative L cells can be blocked by mAb 25-9-17. YAe mAb was used as negative control. (C) D10 responds well to stimulation with Ab presented by spleen cells from normal B6 and Ii-deficient animals. Nonstimulated D10 cells incorporated less than 200 cpm.
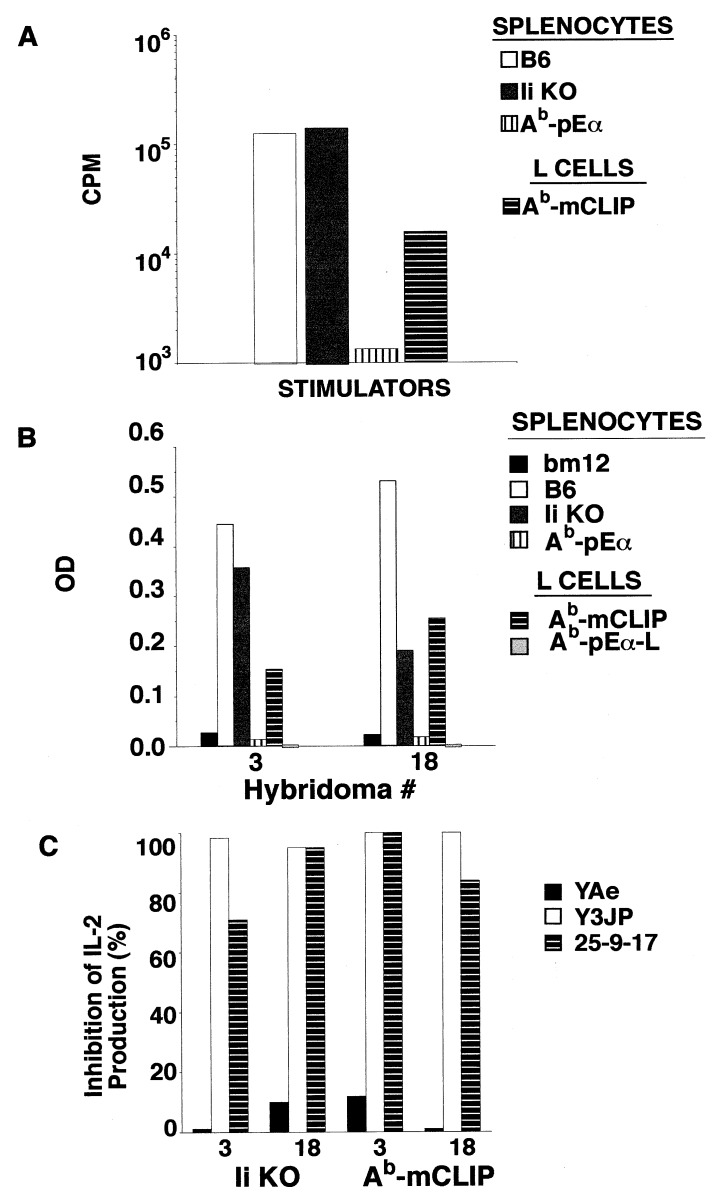
To confirm that the recognition of peptide-dependent conformation was not unique to D10 T cell line, we stimulated purified CD4 cells from bm12 mice in vitro with irradiated splenocytes from Ii KO B6 mice. After two additional restimulations, one of such polyclonal lines was probed for reactivity to Ii KO splenocytes and Ab-mCLIP-expressing L cells. Reactivity to Ab-mCLIP was readily detectable (Fig. 5A). We then fused this cell line with BW5147 T lymphoma to make T cell hybrids to enable analysis at the clonal level. Of 20 hybrids analyzed (with reactivity to Ii KO splenocytes), 8 were reactive to Ab-mCLIP-expressing L cells. The pattern of reactivity of such hybrids was remarkably similar to D10’s (shown for clone 3 and 18 in Fig. 5B), and reactivity of these clones to both Ab-mCLIP-transfected L cells and Ii KO splenocytes could be blocked by mAb 25-9-17 (Fig. 5C). Apparently, the generation of alloreactive T cells with a D10-like recognition pattern is easily achievable.
Figure 5.
T cell recognition of allogeneic MHC class II is not restricted to a single MHC-peptide complex and can be blocked by conformation-sensitive antibodies. (A) Proliferation of a polyclonal T cell line derived from in vitro immunization of bm12 CD4 T cells with Ab+, Ii− splenocytes. Significant proportion of cells reacts to L cells expressing Ab-mCLIP. (B) Recognition pattern of hybridomas (bars 3 and 18 serve as examples) derived from the T cell line shown in A is similar to the allorecognition pattern of D10. Proliferation of IL-2 sensitive CTLL line was measured by Alamar blue colorimetric assay. Results are expressed as arbitrary OD units at 570 nm vs. 600 nm. (C) Conformation-sensitive mAb 25-9-17 blocks the reactivities of the two hybridomas to both Ab+, Ii− splenocytes and Ab-mCLIP covalent complex expressed by Ii− L cell transfectants.
DISCUSSION
Peptide binding is critical for delivery to the cell surface and survival of both MHC class I and II molecules. MHC class I molecules were found to acquire different conformations when different peptides were bound. Those changes were reflected by changes in the binding of anti-class I mAb (12–15). Crystallization of MHC class I with different peptides enabled the detailed analysis of such changes (16, 17). In the present study, we took advantage of the technique allowing MHC class II molecules to be expressed with a single covalently bound peptide and searched for mAb whose interaction with MHC class II would depend upon the nature of the bound peptide.
Conformational transition of MHC class II molecules, which occurs during biosynthesis and trafficking to the cell surface, depends on binding of CLIP and its subsequent DM-catalyzed pH-dependent exchange for another peptide in an endosomal compartment. Acquisition of SDS stability of MHC class II heterodimers is a hallmark of such transition. SDS-stable peptide-bound dimers appeared to be quite heterogeneous as determined by Western blotting with the mAb Y3JP. mAb YAe, which reacts with a known set of MHC–peptide complexes (34), recognized the most “tight” of such dimers, indicating that the nature of a bound peptide may determine the subtle differences in the folding of the MHC class II–peptide complexes. mAb 25-9-17 was shown to interact with compact dimers isolated from I-Ab+ cells but failed to interact with “tight” Ab-pEα complexes recognized by YAe. Blocking of T cell recognition with mAbs revealed that 25-9-17+ and 25-9-17− class II molecules present distinct and diverse sets of peptides.
Because the MHC class II conformations affect T cell reactivity, it is extremely important to understand the molecular basis for these subtle structural transitions. The epitope for 25-9-17 has been previously characterized as being dependent on amino acids β65–67 (Pro-Glu-Ile) and β70 (Arg). Pro is known to break α-helices, and the Pro-Glu-Ile sequence is localized to a “kink ” in the α-helix of the β chain, which may be prone to increased mobility. This kink region has been shown to (i) demonstrate the biggest folding differences for CLIP- and peptide-bound MHC molecules (35, 36); (ii) be critical for a T cell clone (D10) recognition of I-Ab (37), and (iii) be involved in susceptibility to the autoimmune disorder rheumatoid arthritis (36, 38). In the latter case, a residue at position 71 of the HLA-DRβ chain determined the binding of peptides dependent on residues at positions p4 and p6 of the peptide (36, 39, 40). Such binding will most certainly change the conformation of the whole MHC class II molecule, and the kink region in particular. Though the number of the MHC–peptide complexes shown to be recognized by 25-9-17 is limited, it seems that peptides with Pro in position p4 and/or p6 (p1 being represented by Trp, Phe, Tyr, or Met) would promote the 25-9-17+ conformation (see Table 1). The length of peptides is obviously not a factor in determining recognition by 25-9-17 because this mAb reacts with CLIP-bound (19 and 21 amino acids in H2-M knock-out and mCLIP, respectively) and β2-microglobulin peptide-bound (11 amino acids) I-Ab molecules.
The described peptide-dependent conformational transitions of the MHC class II molecules proved to be important for T cell activation. The D10 T cell clone was alloreactive to I-Ab expressed by B6 splenocytes, by Ii knock-out splenocytes, and by L cells as a covalent complex with the Ii fragment CLIP. The same T cells failed to recognize Ab–pEα complex. Similar recognition pattern was found in T cell hybridomas generated from Ab-alloreactive T cells derived from bm12 CD4+ T cells immunized in vitro with Ii-negative spleen cells. mAb 25-9-17 blocked the reactions of either D10 and bm12-anti Ii KO (shown for clones 3 and 18) to Ii-negative cells (L cells and Ii KO splenocytes) and to Ab-mCLIP complex (Table 1 and Figs. 4B and 5C). Thus, multiple MHC class II–peptide complexes can activate T cell alloreactivity and the recognition correlates with the conformation of the MHC–peptide complex detected by mAb.
It seems reasonable to ask whether the thymic selection of T cells is at least partially based on the recognition of distinct MHC conformations. The data from H-2M knock-out (41–43) and Ab-pEα-transgenic animals (44) have shown that both types of conformations (25-9-17+ and 25-9-17−) may mediate positive selection of CD4 T cells. Although negative selection of high-affinity T cells by 25-9-17-negative “tight” C dimers was extremely efficient in Ab-pEα-transgenic animals (no reactivity to Ab-pEα was found in the periphery) the peripheral T cells from these mice were autoreactive to Ab. The Ab-specific reactivity of hybridomas derived from these mice can be blocked by mAb 25-9-17 (data not shown). Thus, T cells with low affinity to positively selecting 25-9-17-negative dimers may potentially be activated in the periphery by autologous peptide–MHC complexes. On the other hand, if the ability to form compact dimers is affected [as in the case of I-Ag7 class II molecule in autoimmune prone non-obese diabetic mice (45)], negative selection of T cells recognizing such dimers is diminished leading to autoimmunity. Thus, it is possible that the balance between “loose” and “tight” conformations expressed during thymic selection determines the presence of self-reactive cells in the periphery. An important example of expression of the “conformationally biased” class II molecules has been published by Dessen et al. (36). This study showed that as a result of a salt bridge formation between Lys-β71 and Asp in position p4 of the bound peptide, the DR4–peptide complex acquired a conformation that was distinct from DR1–HA and especially from DR3–CLIP complexes. Superposition of the three complexes revealed the most significant differences between DR4–collagen peptide (more “tight”) and DR3–CLIP (more “loose”) to be localized to the “kink” part of the β chain α-helix. These data are in line with our hypothesis that the peptide-dependent conformation of the MHC class II molecules affects T cell selection and may lead to autoimmunity when the peptide repertoire is restricted. Restriction of the peptide repertoire or alterations in MHC molecules leading to prevalence of one conformational state expressed during thymic selection may lead to autoimmune recognition in the periphery.
Because MHC–CLIP complexes were found to be in 25-9-17+ conformation, it is important to establish whether such complexes have higher affinity to H2-M molecules than the 25-9-17-negative MHC–peptide complexes. If so, the mechanism of H2-M-mediated transition would become clear. This and the role of MHC class II conformants in positive selection are the matters of current investigation.
Acknowledgments
We thank Peter Cresswell, Andrea J. Sant, Derry Roopenian, and Soon-Cheol Hong for discussions. We also thank Jim Miller, Philippa Marrack, and Luc Van Kaer for the gift of plasmids and mice and Richard Flavell and Idit Shachar for invariant chain knock-out mice. This work was supported in part by National Institutes of Health Grant AI-14579 to C.A.J., Jr. and by the Howard Hughes Medical Institute.
ABBREVIATIONS
- MHC
major histocompatibility complex
- Ii
invariant chain
- CLIP
class II-associated invariant chain peptides
- IL-2
interleukin 2
- FACS
flow cytometry
- WT
wild type
- m
murine
- h
human
References
- 1.Riberdy J M, Newcomb J R, Surman M J, Barbosa J A, Cresswell P. Nature (London) 1992;360:474–477. doi: 10.1038/360474a0. [DOI] [PubMed] [Google Scholar]
- 2.Peterson M, Miller J. Nature (London) 1990;345:172–174. doi: 10.1038/345172a0. [DOI] [PubMed] [Google Scholar]
- 3.Mellins E, Smith L, Arp B, Cotner T, Celis E, Pious D. Nature (London) 1990;343:71–74. doi: 10.1038/343071a0. [DOI] [PubMed] [Google Scholar]
- 4.Germain R N, Hendrix L R. Nature (London) 1991;353:134–139. doi: 10.1038/353134a0. [DOI] [PubMed] [Google Scholar]
- 5.Boniface J J, Lyons D S, Wettstein D A, Allbritton N L, Davis M M. J Exp Med. 1996;183:119–126. doi: 10.1084/jem.183.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Runnels H A, Mooreand J C, Jensen P E. J Exp Med. 1996;183:127–136. doi: 10.1084/jem.183.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen P E. J Exp Med. 1990;171:1779–1784. doi: 10.1084/jem.171.5.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen P E. J Exp Med. 1991;174:1111–1120. doi: 10.1084/jem.174.5.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadegh-Nasseri S, Germain R N. Nature (London) 1991;353:167–170. doi: 10.1038/353167a0. [DOI] [PubMed] [Google Scholar]
- 10.Denzin L K, Cresswell P. Cell. 1995;82:155–165. doi: 10.1016/0092-8674(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 11.Sloan V S, Cameron P, Porter G, Gammon M, Amaya M, Mellins E, Zaller D M. Nature (London) 1995;375:802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- 12.Bluestone J A, Jameson S, Miller S, Dick R, II. J Exp Med. 1992;176:1757–1761. doi: 10.1084/jem.176.6.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherman L A, Chattopadhyay S, Biggs J A, Dick R F, II, Bluestone J A. Proc Natl Acad Sci USA. 1993;90:6949–6951. doi: 10.1073/pnas.90.15.6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pullen J K, Tallquist M D, Melvold R W, Pease L R. J Immunol. 1994;152:3445–3452. [PubMed] [Google Scholar]
- 15.Rohren E M, McCormick D J, Pease L R. J Immunol. 1994;152:5337–5343. [PubMed] [Google Scholar]
- 16.Fremont D H, Matsumura M, Stura E A, Peterson P A, Wilson I A. Science. 1992;257:919–927. doi: 10.1126/science.1323877. [DOI] [PubMed] [Google Scholar]
- 17.Stura E A, Matsumura M, Fremont D H, Saito Y, Peterson P A, Wilson I A. J Mol Biol. 1992;228:975–982. doi: 10.1016/0022-2836(92)90881-j. [DOI] [PubMed] [Google Scholar]
- 18.Stebbins C C, Loss G E, Jr, Elias C G, Chervonsky A, Sant A J. J Exp Med. 1995;181:223–234. doi: 10.1084/jem.181.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davidson R L, Gerald P A. Somatic Cell Genet. 1976;2:165–176. doi: 10.1007/BF01542629. [DOI] [PubMed] [Google Scholar]
- 20.Kaye J, Janeway C A., Jr J Exp Med. 1984;159:1397–1412. doi: 10.1084/jem.159.5.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudensky A Y, Rath S, Preston-Hurlburt P, Murphy D, Janeway C A., Jr Nature (London) 1991;353:660–662. doi: 10.1038/353660a0. [DOI] [PubMed] [Google Scholar]
- 22.Ozato K, Mayer N, Sachs D H. J Immunol. 1980;124:533–540. [PubMed] [Google Scholar]
- 23.Bhattacharya A, Dorf M E, Springer T A. J Immunol. 1981;127:2488–2495. [PubMed] [Google Scholar]
- 24.Ledbetter J A, Herzenberg L A. Immunol Rev. 1979;47:63–90. doi: 10.1111/j.1600-065x.1979.tb00289.x. [DOI] [PubMed] [Google Scholar]
- 25.Janeway C A, Jr, Conrad P S, Lerner E A, Babich J, Wettstein P, Murphy D B. J Immunol. 1984;132:662–667. [PubMed] [Google Scholar]
- 26.Murphy D B, Lo D, Rath S, Brinster R L, Flavell R A, Slanetz A, Janeway C A., Jr Nature (London) 1989;338:765–768. doi: 10.1038/338765a0. [DOI] [PubMed] [Google Scholar]
- 27.Denzin L K, Robbins N F, Carboy-Newcomb C, Cresswell P. Immunity. 1994;1:595–606. doi: 10.1016/1074-7613(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 28.Sant A J, Braunstein N S, Germain R N. Proc Natl Acad Sci USA. 1987;84:8065–8069. doi: 10.1073/pnas.84.22.8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chervonsky A V, Gordon L, Sant A J. Int Immunol. 1994;6:973–982. doi: 10.1093/intimm/6.7.973. [DOI] [PubMed] [Google Scholar]
- 30.Rudensky A Y, Preston-Hurlburt P, Hong S-C, Barlow A, Janeway C A., Jr Nature (London) 1991;353:622–624. doi: 10.1038/353622a0. [DOI] [PubMed] [Google Scholar]
- 31.Ignatowicz L, Winslow G, Bill J, Kappler J, Marrack P. J Immunol. 1995;154:3852–3862. [PubMed] [Google Scholar]
- 32.Sette A, Ceman S, Kubo R T, Sakaguchi K, Appella E, Hunt D F, Davis T A, Michel H, Shabanowitz J, Rudersdorf R, Grey H M, Demars R. Science. 1992;258:1801–1804. doi: 10.1126/science.1465617. [DOI] [PubMed] [Google Scholar]
- 33.Grubin C E, Kovats S, deRoos P, Rudensky A Y. Immunity. 1997;7:197–208. doi: 10.1016/s1074-7613(00)80523-3. [DOI] [PubMed] [Google Scholar]
- 34.Rudensky A Y, Preston-Hurlburt P, al-Ramadi B K, Rothbard J, Janeway C A., Jr Nature (London) 1992;269:429–431. doi: 10.1038/359429a0. [DOI] [PubMed] [Google Scholar]
- 35.Ghosh P, Amaya M, Mellins E, Wiley D C. Nature (London) 1995;378:457–462. doi: 10.1038/378457a0. [DOI] [PubMed] [Google Scholar]
- 36.Dessen A, Lawrence C M, Cupo S, Zaller D M, Wiley D C. Immunity. 1997;7:473–481. doi: 10.1016/s1074-7613(00)80369-6. [DOI] [PubMed] [Google Scholar]
- 37.Hong S C, Chelouche A, Lin R H, Shaywitz D, Braunstein N S, Glimcher L, Janeway C A., Jr Cell. 1992;69:999–1009. doi: 10.1016/0092-8674(92)90618-m. [DOI] [PubMed] [Google Scholar]
- 38.Gregersen P K, Silver J, Winchester R J. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 39.Woulfe S L, Bono C P, Zacheis D A, Kirschmann T A, Baudino C S, Karr R W, Schwartz B D. Arthritis Rheum. 1995;38:1744–1753. doi: 10.1002/art.1780381207. [DOI] [PubMed] [Google Scholar]
- 40.Hammer J, Gallazzi F, Bono E, Karr R W, Guenot J, Valsasnini P, Nagy Z A, Sinigaglia F. J Exp Med. 1995;181:1847–1855. doi: 10.1084/jem.181.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyazaki T, Wolf P, Tourne S, Waltzinger C, Dierich A, Barois N, Ploegh H, Benoist C, Mathis D. Cell. 1996;84:531–541. doi: 10.1016/s0092-8674(00)81029-6. [DOI] [PubMed] [Google Scholar]
- 42.Martin W D, Hicks G G, Mendiratta S K, Leva H I, Ruley H E, Van Kaer L. Cell. 1996;84:543–550. doi: 10.1016/s0092-8674(00)81030-2. [DOI] [PubMed] [Google Scholar]
- 43.Fung-Leung W P, Surh C D, Liljedahl M, Pang J, Leturcq D, Peterson P A, Webb S, Karlsson L. Science. 1996;271:1278–1281. doi: 10.1126/science.271.5253.1278. [DOI] [PubMed] [Google Scholar]
- 44.Ignatowicz L, Kappler J, Marrack P. Cell. 1996;84:521–529. doi: 10.1016/s0092-8674(00)81028-4. [DOI] [PubMed] [Google Scholar]
- 45.Carrasco-Marin E, Shimizu J, Kanagawa O, Unanue E R. J Immunol. 1996;156:450–458. [PubMed] [Google Scholar]