Abstract
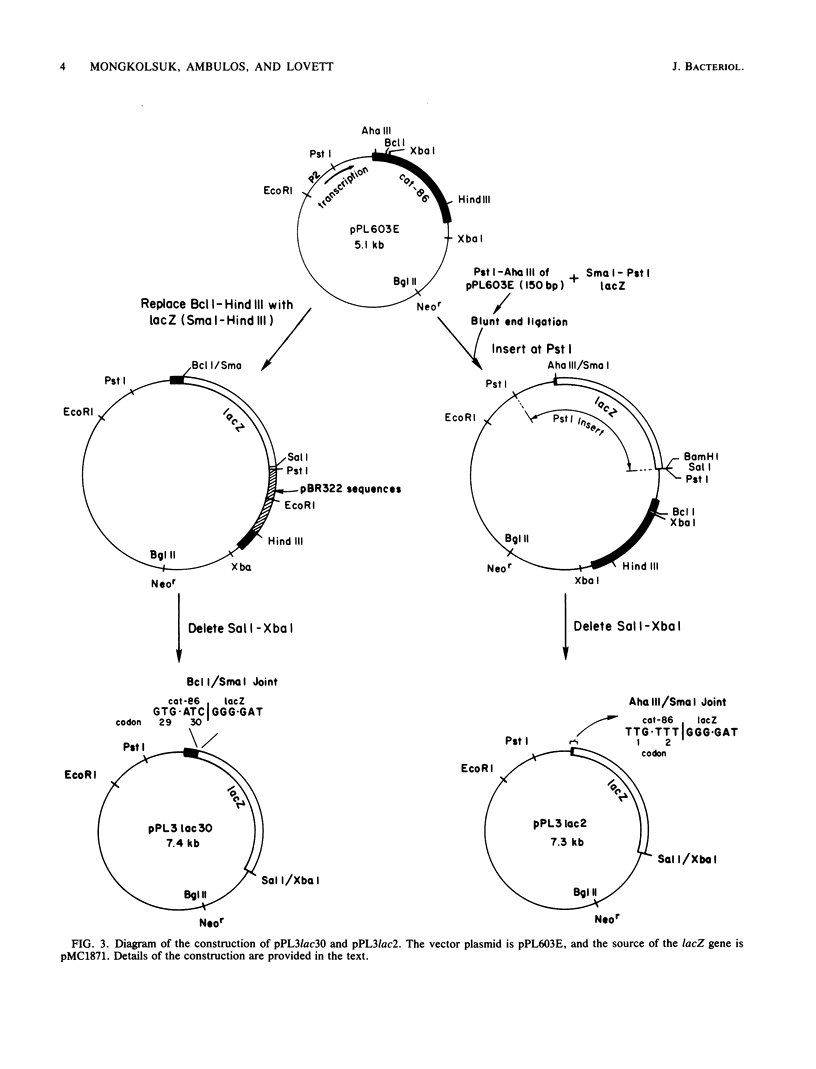
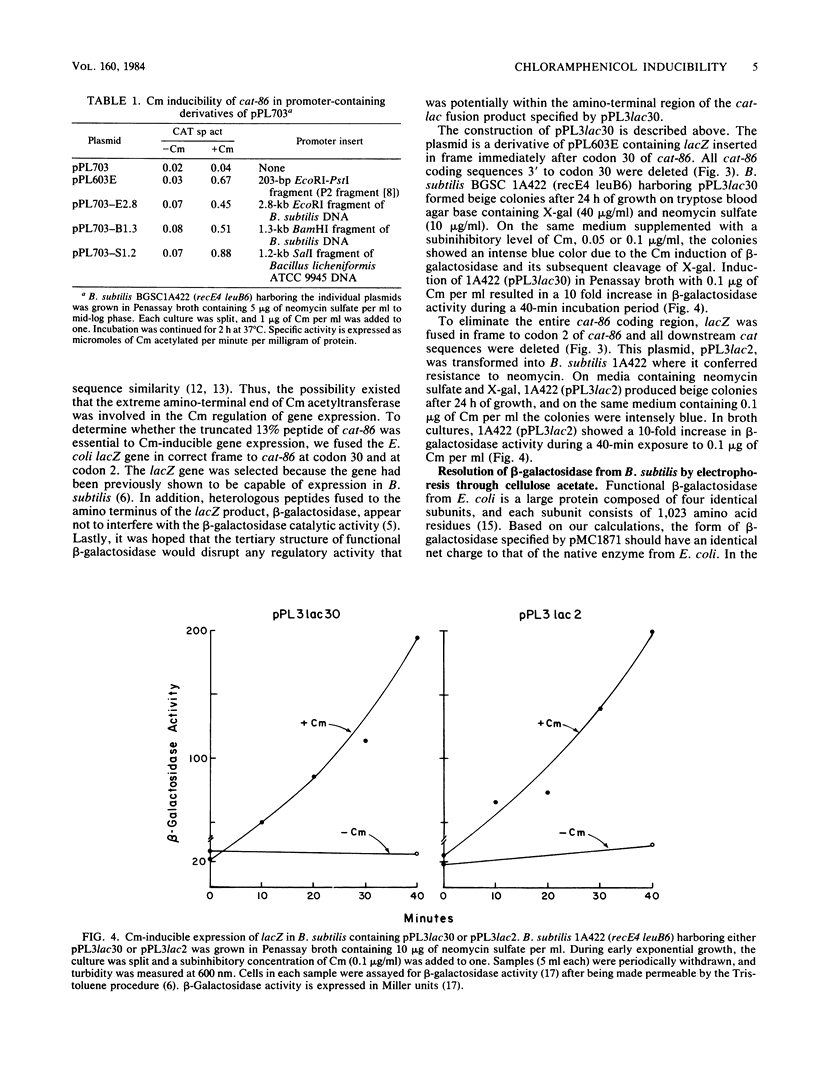
cat-86 specifies chloramphenicol acetyltransferase and is the indicator gene on the Bacillus subtilis promoter cloning plasmid pPL703. Insertion of promoters from various sources into pPL703 at a site ca. 144 base pairs upstream from cat-86 activates expression of cat-86, and the expression is characteristically inducible by chloramphenicol. Thus, chloramphenicol inducibility of cat-86 is independent of the promoter that is used to activate the gene. To determine whether cat-86 or its products were involved in chloramphenicol inducibility, gene replacement studies were performed. cat-86 consists of 220 codons. The lacZ gene from Escherichia coli was inserted into a promoter-containing derivative of pPL703, plasmid pPL603E, at two locations within cat-86. pPL3lac2 contains lacZ inserted in frame after codon 2 of cat-86. pPL3lac30 contains lacZ inserted in frame after codon 30 of cat-86. In both constructions, all cat coding sequences 3' to the site of the lacZ insertion were deleted. Both plasmids exhibited chloramphenicol inducibility of beta-galactosidase in B. subtilis. These studies provide the first direct demonstration that the transcription and translation products of a chloramphenicol-inducible cat gene are uninvolved in chloramphenicol inducibility of gene expression. The results localize the region essential to inducibility to the 144-base pair segment that intervenes between the site of promoter insertion and the cat-86 gene.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambulos N. P., Jr, Chow J. H., Mongkolsuk S., Preis L. H., Vollmar W. R., 2nd, Lovett P. S. Constitutive variants of the pC194 cat gene exhibit DNA alterations in the vicinity of the ribosome binding site sequence. Gene. 1984 May;28(2):171–176. doi: 10.1016/0378-1119(84)90254-3. [DOI] [PubMed] [Google Scholar]
- Bott K. F., Wilson G. A. Development of competence in the Bacillus subtilis transformation system. J Bacteriol. 1967 Sep;94(3):562–570. doi: 10.1128/jb.94.3.562-570.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Byeon W. H., Weisblum B. Post-transcriptional regulation of chloramphenicol acetyl transferase. J Bacteriol. 1984 May;158(2):543–550. doi: 10.1128/jb.158.2.543-550.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Martinez-Arias A., Shapira S. K., Chou J. Beta-galactosidase gene fusions for analyzing gene expression in escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- Donnelly C. E., Sonenshein A. L. Promoter-probe plasmid for Bacillus subtilis. J Bacteriol. 1984 Mar;157(3):965–967. doi: 10.1128/jb.157.3.965-967.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall E. J., Williams D. M., Lovett P. S., Rudolph C., Vasantha N., Guyer M. Chloramphenicol-inducible gene expression in Bacillus subtilis. Gene. 1983 Oct;24(2-3):171–177. doi: 10.1016/0378-1119(83)90077-x. [DOI] [PubMed] [Google Scholar]
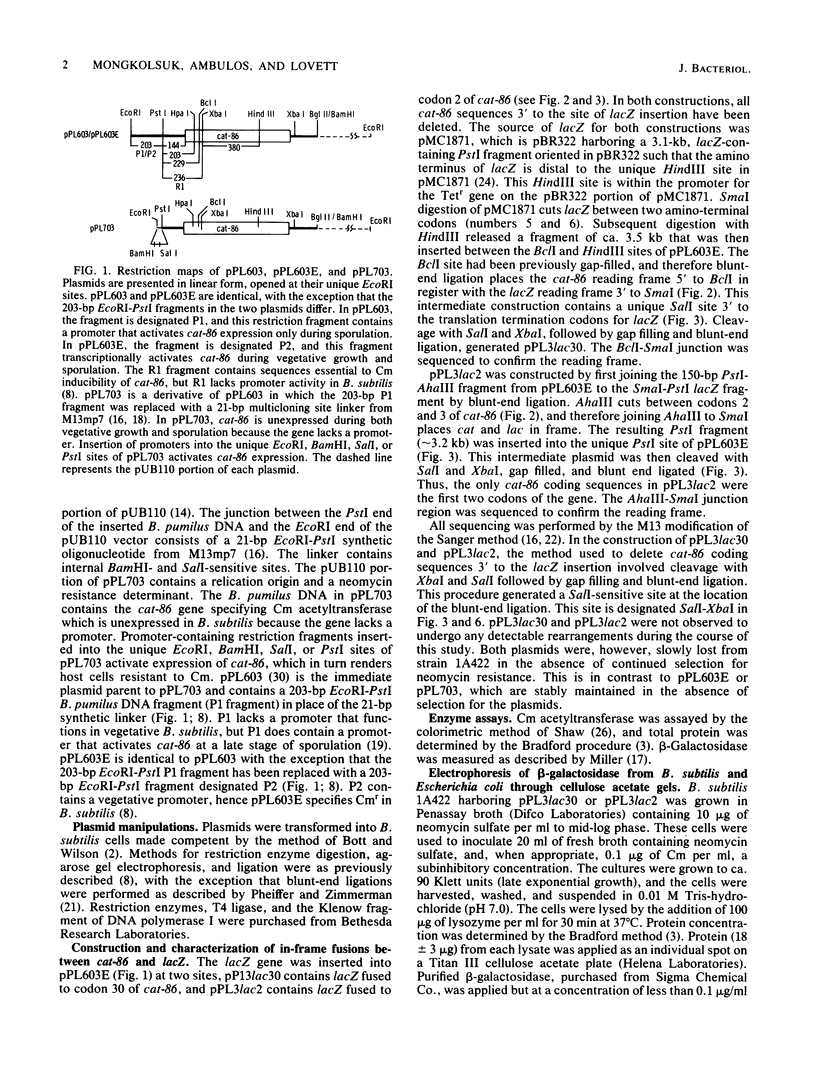
- Duvall E. J., Williams D. M., Mongkolsuk S., Lovett P. S. Regulatory regions that control expression of two chloramphenicol-inducible cat genes cloned in Bacillus subtilis. J Bacteriol. 1984 Jun;158(3):784–790. doi: 10.1128/jb.158.3.784-790.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich S. D. Replication and expression of plasmids from Staphylococcus aureus in Bacillus subtilis. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1680–1682. doi: 10.1073/pnas.74.4.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicher E. M., Washburn L. L. Assignment of genes to regions of mouse chromosomes. Proc Natl Acad Sci U S A. 1978 Feb;75(2):946–950. doi: 10.1073/pnas.75.2.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R., Yanofsky C. A complementary DNA oligomer releases a transcription pause complex. J Biol Chem. 1983 Aug 10;258(15):9208–9212. [PubMed] [Google Scholar]
- Harwood C. R., Williams D. M., Lovett P. S. Nucleotide sequence of a Bacillus pumilus gene specifying chloramphenicol acetyltransferase. Gene. 1983 Oct;24(2-3):163–169. doi: 10.1016/0378-1119(83)90076-8. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalanko A., Palva I., Söderlund Restriction maps of plasmids pUB110 and pBD9. Gene. 1981 Sep;14(4):325–328. doi: 10.1016/0378-1119(81)90165-7. [DOI] [PubMed] [Google Scholar]
- Kalnins A., Otto K., Rüther U., Müller-Hill B. Sequence of the lacZ gene of Escherichia coli. EMBO J. 1983;2(4):593–597. doi: 10.1002/j.1460-2075.1983.tb01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsuk S., Chiang Y. W., Reynolds R. B., Lovett P. S. Restriction fragments that exert promoter activity during postexponential growth of Bacillus subtilis. J Bacteriol. 1983 Sep;155(3):1399–1406. doi: 10.1128/jb.155.3.1399-1406.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsuk S., Lovett P. S. Selective expression of a plasmid cat gene at a late stage of Bacillus subtilis sporulation. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3457–3460. doi: 10.1073/pnas.81.11.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pheiffer B. H., Zimmerman S. B. Polymer-stimulated ligation: enhanced blunt- or cohesive-end ligation of DNA or deoxyribooligonucleotides by T4 DNA ligase in polymer solutions. Nucleic Acids Res. 1983 Nov 25;11(22):7853–7871. doi: 10.1093/nar/11.22.7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoner R. G., Williams D. M., Lovett P. S. Enhanced expression of mouse dihydrofolate reductase in Bacillus subtilis. Gene. 1983 Apr;22(1):47–57. doi: 10.1016/0378-1119(83)90063-x. [DOI] [PubMed] [Google Scholar]
- Shapira S. K., Chou J., Richaud F. V., Casadaban M. J. New versatile plasmid vectors for expression of hybrid proteins coded by a cloned gene fused to lacZ gene sequences encoding an enzymatically active carboxy-terminal portion of beta-galactosidase. Gene. 1983 Nov;25(1):71–82. doi: 10.1016/0378-1119(83)90169-5. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase: enzymology and molecular biology. CRC Crit Rev Biochem. 1983;14(1):1–46. doi: 10.3109/10409238309102789. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Williams D. M., Duvall E. J., Lovett P. S. Cloning restriction fragments that promote expression of a gene in Bacillus subtilis. J Bacteriol. 1981 Jun;146(3):1162–1165. doi: 10.1128/jb.146.3.1162-1165.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. M., Schoner R. G., Duvall E. J., Preis L. H., Lovett P. S. Expression of Escherichia coli trp genes and the mouse dihydrofolate reductase gene cloned in Bacillus subtilis. Gene. 1981 Dec;16(1-3):199–206. doi: 10.1016/0378-1119(81)90076-7. [DOI] [PubMed] [Google Scholar]
- Wilson C. R., Skinner S. E., Shaw W. V. Analysis of two chloramphenicol resistance plasmids from Staphylococcus aureus: insertional inactivation of Cm resistance, mapping of restriction sites, and construction of cloning vehicles. Plasmid. 1981 May;5(3):245–258. doi: 10.1016/0147-619x(81)90002-0. [DOI] [PubMed] [Google Scholar]
- Winshell E., Shaw W. V. Kinetics of induction and purification of chloramphenicol acetyltransferase from chloramphenicol-resistant Staphylococcus aureus. J Bacteriol. 1969 Jun;98(3):1248–1257. doi: 10.1128/jb.98.3.1248-1257.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zukowski M. M., Gaffney D. F., Speck D., Kauffmann M., Findeli A., Wisecup A., Lecocq J. P. Chromogenic identification of genetic regulatory signals in Bacillus subtilis based on expression of a cloned Pseudomonas gene. Proc Natl Acad Sci U S A. 1983 Feb;80(4):1101–1105. doi: 10.1073/pnas.80.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]