Abstract
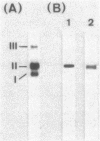
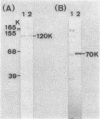
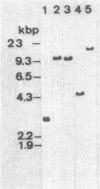
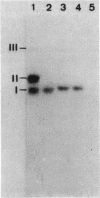
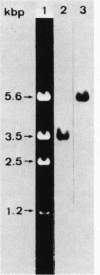
Bacillus stearothermophilus IAM11001 produced three beta-galactosidases, beta-galactosidase I, II, and III (beta-gal I, II, and III), which are detectable by polyacrylamide (nondenatured) gel electrophoresis. By connecting restriction fragments of the chromosomal DNA to plasmid vectors, followed by transformation of Escherichia coli, two beta-galactosidase genes (bgaA and bgaB) located close to each other on the chromosome were isolated. Identification of the gene products and Southern hybridization analyses with a 2.7-kilobase-pair EcoRI fragment containing the bgaA gene as probe revealed that a single bgaA gene exists on the genome and that beta-gal II and beta-gal III consist of a common subunit (the bgaA gene product; molecular weight, 120,000), but differ in their assembly (beta-gal II is a dimer, and beta-gal III is a tetramer). The bgaB gene product (molecular weight, 70,000) in Bacillus subtilis harboring pHG5 (a hybrid plasmid consisting of pUB110 and a 2.9-kilobase-pair EcoRI fragment) was estimated to be the beta-gal I protein from its heat stability. Southern hybridization and immunological testing indicated that the two genes have no homology.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Backman K., Ptashne M., Gilbert W. Construction of plasmids carrying the cI gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4174–4178. doi: 10.1073/pnas.73.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- CRAVEN G. R., STEERS E., Jr, ANFINSEN C. B. PURIFICATION, COMPOSITION, AND MOLECULAR WEIGHT OF THE BETA-GALACTOSIDASE OF ESCHERICHIA COLI K12. J Biol Chem. 1965 Jun;240:2468–2477. [PubMed] [Google Scholar]
- Campbell J. H., Lengyel J. A., Langridge J. Evolution of a second gene for beta-galactosidase in Escherichia coli. Proc Natl Acad Sci U S A. 1973 Jun;70(6):1841–1845. doi: 10.1073/pnas.70.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Cohen S. N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979 Jan 5;168(1):111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson R. P., Steers E., Jr Comparative study of isoenzyme formation of bacterial beta-galactosidase. J Bacteriol. 1970 Apr;102(1):79–84. doi: 10.1128/jb.102.1.79-84.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler A. V., Zabin I. Amino acid sequence of beta-galactosidase. XI. Peptide ordering procedures and the complete sequence. J Biol Chem. 1978 Aug 10;253(15):5521–5525. [PubMed] [Google Scholar]
- Goodman R. E., Pederson D. M. beta-Galactosidase from Bacillus stearothermophilus. Can J Microbiol. 1976 Jun;22(6):817–825. doi: 10.1139/m76-118. [DOI] [PubMed] [Google Scholar]
- Griffiths M. W., Muir D. D. Properties of a thermostable beta-galactosidase from a thermophilic Bacillus: comparison of the enzyme activity of whole cells, purified enzyme and immobilised whole cells. J Sci Food Agric. 1978 Sep;29(9):753–761. doi: 10.1002/jsfa.2740290904. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Contente S., Dubnau D. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J Bacteriol. 1978 Apr;134(1):318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G. Experimental evolution of a new enzymatic function. Kinetic analysis of the ancestral (ebg) and evolved (ebg) enzymes. J Mol Biol. 1976 Oct 15;107(1):71–84. doi: 10.1016/s0022-2836(76)80018-6. [DOI] [PubMed] [Google Scholar]
- Hall B. G., Reeve E. C. A third beta-galactosidase in a strain of Klebsiella that possesses two lac genes. J Bacteriol. 1977 Oct;132(1):219–223. doi: 10.1128/jb.132.1.219-223.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick J. L., Smith A. J. Size and charge isomer separation and estimation of molecular weights of proteins by disc gel electrophoresis. Arch Biochem Biophys. 1968 Jul;126(1):155–164. doi: 10.1016/0003-9861(68)90569-9. [DOI] [PubMed] [Google Scholar]
- Kemp D. J., Cowman A. F. Detection of expressed polypeptides by direct immunoassay of colonies. Methods Enzymol. 1981;79(Pt B):622–630. doi: 10.1016/s0076-6879(81)79087-6. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Negoro S., Shinagawa H., Nakata A., Kinoshita S., Hatozaki T., Okada H. Plasmid control of 6-aminohexanoic acid cyclic dimer degradation enzymes of Flavobacterium sp. KI72. J Bacteriol. 1980 Jul;143(1):238–245. doi: 10.1128/jb.143.1.238-245.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negoro S., Taniguchi T., Kanaoka M., Kimura H., Okada H. Plasmid-determined enzymatic degradation of nylon oligomers. J Bacteriol. 1983 Jul;155(1):22–31. doi: 10.1128/jb.155.1.22-31.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- SAITO H., MIURA K. I. PREPARATION OF TRANSFORMING DEOXYRIBONUCLEIC ACID BY PHENOL TREATMENT. Biochim Biophys Acta. 1963 Aug 20;72:619–629. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich J. T., McFeters G. A., Temple K. L. Induction and characterization of -galactosidase in an extreme thermophile. J Bacteriol. 1972 May;110(2):691–698. doi: 10.1128/jb.110.2.691-698.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]