Abstract
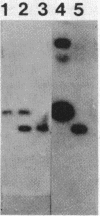
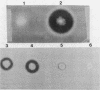
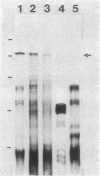
The neutral protease gene of Bacillus subtilis has been cloned, and its nucleotide sequence has been determined. The cloned gene was used to create an in vitro-derived deletion mutation, which was used to replace the wild-type copy of the gene. This deletion, in combination with a deletion of the alkaline protease gene, completely abolished protease production. The loss of the proteases had no detectable effect on growth, morphology, or sporulation.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman K., Ptashne M., Gilbert W. Construction of plasmids carrying the cI gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4174–4178. doi: 10.1073/pnas.73.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band L., Henner D. J. Bacillus subtilis requires a "stringent" Shine-Dalgarno region for gene expression. DNA. 1984;3(1):17–21. doi: 10.1089/dna.1.1984.3.17. [DOI] [PubMed] [Google Scholar]
- Band L., Shimotsu H., Henner D. J. Nucleotide sequence of the Bacillus subtilis trpE and trpD genes. Gene. 1984 Jan;27(1):55–65. doi: 10.1016/0378-1119(84)90238-5. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner F. R., Williams B. G., Blechl A. E., Denniston-Thompson K., Faber H. E., Furlong L., Grunwald D. J., Kiefer D. O., Moore D. D., Schumm J. W. Charon phages: safer derivatives of bacteriophage lambda for DNA cloning. Science. 1977 Apr 8;196(4286):161–169. doi: 10.1126/science.847462. [DOI] [PubMed] [Google Scholar]
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978 Oct;4(2):121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Effect of growth conditions on the formation of the relaxation complex of supercoiled ColE1 deoxyribonucleic acid and protein in Escherichia coli. J Bacteriol. 1972 Jun;110(3):1135–1146. doi: 10.1128/jb.110.3.1135-1146.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crea R., Horn T. Synthesis of oligonucleotides on cellulose by a phosphotriester method. Nucleic Acids Res. 1980 May 24;8(10):2331–2348. doi: 10.1093/nar/8.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F. A., Nguyen A., Lang D., Hoch J. A. Construction and properties of an integrable plasmid for Bacillus subtilis. J Bacteriol. 1983 Jun;154(3):1513–1515. doi: 10.1128/jb.154.3.1513-1515.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Contente S., Dubnau D. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J Bacteriol. 1978 Apr;134(1):318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershfield V., Boyer H. W., Yanofsky C., Lovett M. A., Helinski D. R. Plasmid ColEl as a molecular vehicle for cloning and amplification of DNA. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3455–3459. doi: 10.1073/pnas.71.9.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J. A., Barat M., Anagnostopoulos C. Transformation and transduction in recombination-defective mutants of Bacillus subtilis. J Bacteriol. 1967 Jun;93(6):1925–1937. doi: 10.1128/jb.93.6.1925-1937.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J. A. Genetics of bacterial sporulation. Adv Genet. 1976;18:69–98. doi: 10.1016/s0065-2660(08)60437-x. [DOI] [PubMed] [Google Scholar]
- Jolliffe L. K., Doyle R. J., Streips U. N. Extracellular proteases modify cell wall turnover in Bacillus subtilis. J Bacteriol. 1980 Mar;141(3):1199–1208. doi: 10.1128/jb.141.3.1199-1208.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreil G. Transfer of proteins across membranes. Annu Rev Biochem. 1981;50:317–348. doi: 10.1146/annurev.bi.50.070181.001533. [DOI] [PubMed] [Google Scholar]
- Lawn R. M., Adelman J., Bock S. C., Franke A. E., Houck C. M., Najarian R. C., Seeburg P. H., Wion K. L. The sequence of human serum albumin cDNA and its expression in E. coli. Nucleic Acids Res. 1981 Nov 25;9(22):6103–6114. doi: 10.1093/nar/9.22.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton T. J., Doi R. H. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J Biol Chem. 1971 May 25;246(10):3189–3195. [PubMed] [Google Scholar]
- Levy P. L., Pangburn M. K., Burstein Y., Ericsson L. H., Neurath H., Walsh K. A. Evidence of homologous relationship between thermolysin and neutral protease A of Bacillus subtilis. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4341–4345. doi: 10.1073/pnas.72.11.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett P. S., Keggins K. M. Bacillus subtilis as a host for molecular cloning. Methods Enzymol. 1979;68:342–357. doi: 10.1016/0076-6879(79)68025-4. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus beta-lactamase gene. J Biol Chem. 1981 Nov 10;256(21):11283–11291. [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Mills D. R., Kramer F. R. Structure-independent nucleotide sequence analysis. Proc Natl Acad Sci U S A. 1979 May;76(5):2232–2235. doi: 10.1073/pnas.76.5.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Murray C. L., Rabinowitz J. C. Nucleotide sequences of transcription and translation initiation regions in Bacillus phage phi 29 early genes. J Biol Chem. 1982 Jan 25;257(2):1053–1062. [PubMed] [Google Scholar]
- Ohmura K., Shiroza T., Nakamura K., Nakayama A., Yamane K., Yoda K., Yamasaki M., Tamura G. A Bacillus subtilis secretion vector system derived from the B. subtilis alpha-amylase promoter and signal sequence region, and secretion of Escherichia coli beta-lactamase by the vector system. J Biochem. 1984 Jan;95(1):87–93. doi: 10.1093/oxfordjournals.jbchem.a134607. [DOI] [PubMed] [Google Scholar]
- Palva I., Lehtovaara P., Käriäinen L., Sibakov M., Cantell K., Schein C. H., Kashiwagi K., Weissmann C. Secretion of interferon by Bacillus subtilis. Gene. 1983 May-Jun;22(2-3):229–235. doi: 10.1016/0378-1119(83)90107-5. [DOI] [PubMed] [Google Scholar]
- Palva I., Sarvas M., Lehtovaara P., Sibakov M., Käriäinen L. Secretion of Escherichia coli beta-lactamase from Bacillus subtilis by the aid of alpha-amylase signal sequence. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5582–5586. doi: 10.1073/pnas.79.18.5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal M., Kunst F., Lepesant J. A., Dedonder R. Characterization of two sucrase activities in Bacillus subtilis Marburg. Biochimie. 1971;53(10):1059–1066. doi: 10.1016/s0300-9084(71)80193-1. [DOI] [PubMed] [Google Scholar]
- Piggot P. J., Coote J. G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976 Dec;40(4):908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest F. G. Extracellular enzyme synthesis in the genus Bacillus. Bacteriol Rev. 1977 Sep;41(3):711–753. doi: 10.1128/br.41.3.711-753.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitsch C. A., Hageman J. H. Bacillopeptidase F: two forms of a glycoprotein serine protease from Bacillus subtilis 168. J Bacteriol. 1983 Jul;155(1):145–152. doi: 10.1128/jb.155.1.145-152.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoer R., Rappaport H. P. Analysis of a Bacillus subtilis proteinase mutant. J Bacteriol. 1972 Feb;109(2):575–583. doi: 10.1128/jb.109.2.575-583.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M. L., Ferrari E. Replacement of the Bacillus subtilis subtilisin structural gene with an In vitro-derived deletion mutation. J Bacteriol. 1984 May;158(2):411–418. doi: 10.1128/jb.158.2.411-418.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara H., Yamane K., Maruo B. Thermosensitive, extracellular neutral proteases in Bacillus subtilis: isolation, characterization, and genetics. J Bacteriol. 1979 Aug;139(2):583–590. doi: 10.1128/jb.139.2.583-590.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara H., Yoneda Y., Yamane K., Maruo B. Regulation of neutral protease productivity in Bacillus subtilis: transformation of high protease productivity. J Bacteriol. 1974 Jul;119(1):82–91. doi: 10.1128/jb.119.1.82-91.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J. A., Ferrari E., Henner D. J., Estell D. A., Chen E. Y. Cloning, sequencing, and secretion of Bacillus amyloliquefaciens subtilisin in Bacillus subtilis. Nucleic Acids Res. 1983 Nov 25;11(22):7911–7925. doi: 10.1093/nar/11.22.7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Galizzi A., Henner D. Nucleotide sequence of the amylase gene from Bacillus subtilis. Nucleic Acids Res. 1983 Jan 25;11(2):237–249. doi: 10.1093/nar/11.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]