Abstract
Caulobacter crescentus trp mutants were identified from a collection of auxotrophs. Precursor feeding experiments, accumulation studies, and complementation experiments resulted in the identification of six genes corresponding to trpA, trpB, trpC, trpD, trpE, and trpF. Genetic mapping experiments demonstrated that the trp genes were in two clusters, trpCDE and trpFBA, and a 5.4-kilobase restriction fragment from the C. crescentus chromosome was isolated that contained the trpFBA gene cluster. Complementation experiments with clones containing the 5.4-kilobase fragment indicated that trpF was expressed in Escherichia coli and that all three genes were expressed in Pseudomonas putida. This expression was lost in both organisms when the pBR322 tet gene promoter was inactivated, indicating that all three genes were transcribed in the same orientation from the tet promoter. Thus, the C. crescentus promoters do not seem to be expressed in E. coli or P. putida. Complementation of the C. crescentus trp mutants indicated that the tet promoter was not necessary for expression in C. crescentus and suggested that at least two native promoters were present for expression of the trpF, trpB, and trpA genes. Taken together, these results indicate that C. crescentus promoters may have structures that are significantly different from the promoters of other gram-negative species.
Full text
PDF

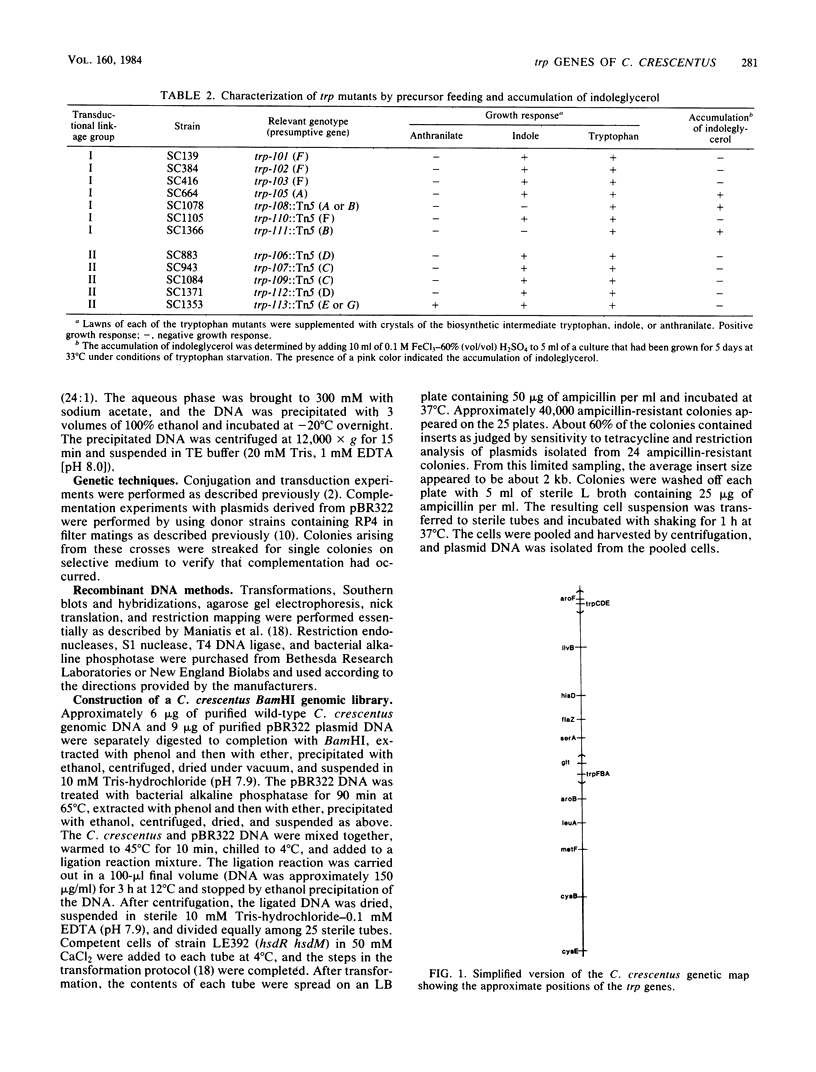
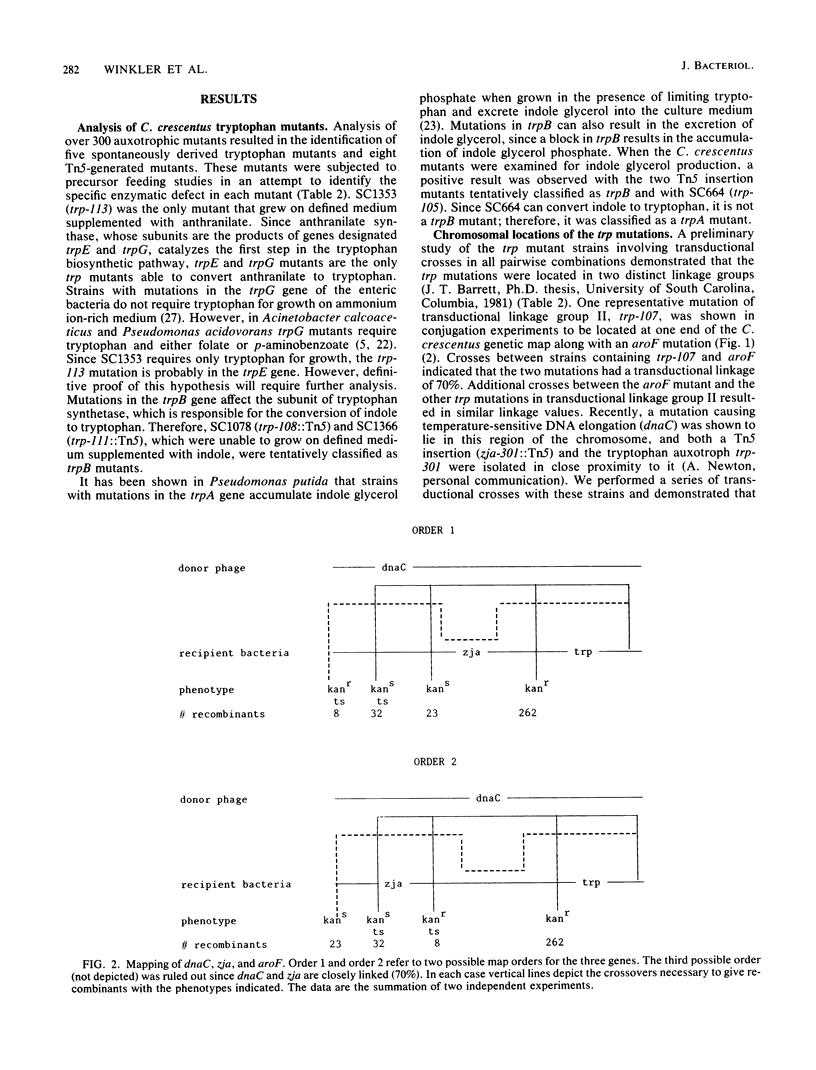


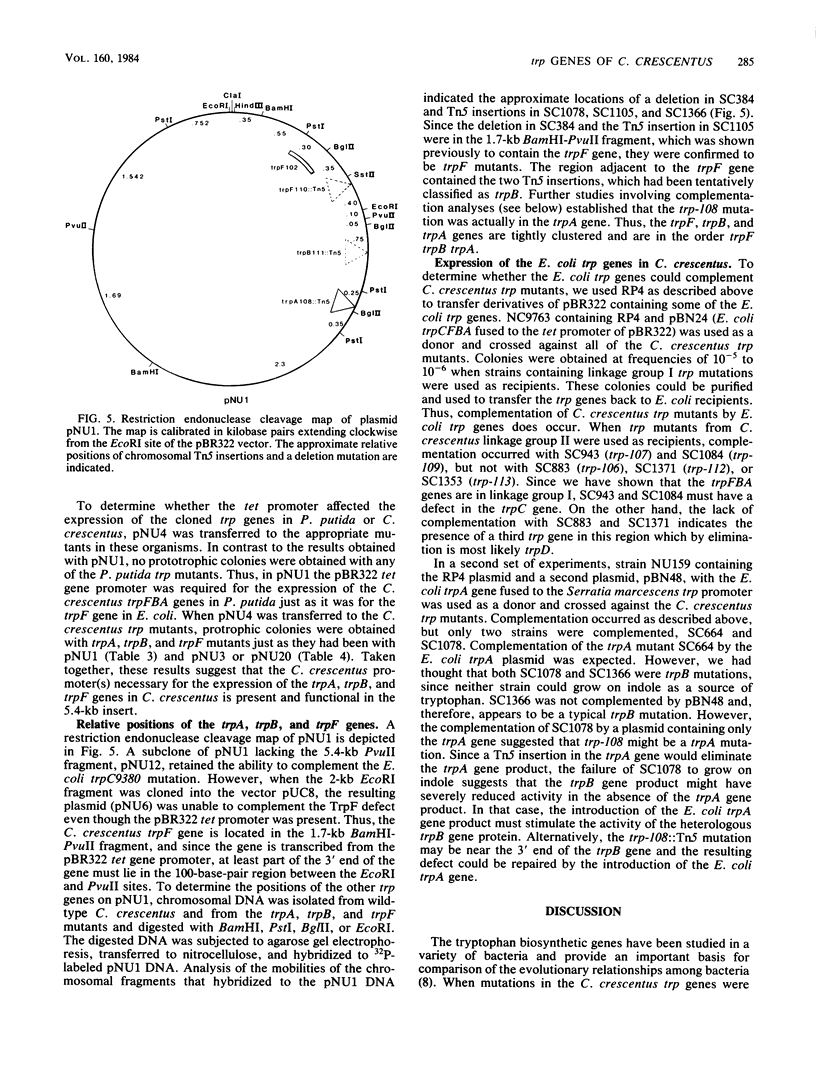


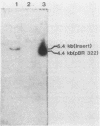
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agabian N., Evinger M., Parker G. Generation of asymmetry during development. Segregation of type-specific proteins in Caulobacter. J Cell Biol. 1979 Apr;81(1):123–136. doi: 10.1083/jcb.81.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya K., Shapiro L. In vitro transcription of the early region of Caulobacter phage phi Cd1 deoxyribonucleic acid by host RNA polymerase. Biochemistry. 1982 Sep 14;21(19):4707–4713. doi: 10.1021/bi00262a029. [DOI] [PubMed] [Google Scholar]
- Barrett J. T., Croft R. H., Ferber D. M., Gerardot C. J., Schoenlein P. V., Ely B. Genetic mapping with Tn5-derived auxotrophs of Caulobacter crescentus. J Bacteriol. 1982 Aug;151(2):888–898. doi: 10.1128/jb.151.2.888-898.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J. T., Rhodes C. S., Ferber D. M., Jenkins B., Kuhl S. A., Ely B. Construction of a genetic map for Caulobacter crescentus. J Bacteriol. 1982 Mar;149(3):889–896. doi: 10.1128/jb.149.3.889-896.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvinger W. E., Stone L. C., Heath H. E. Biochemical genetics of tryptophan synthesis in Pseudomonas acidovorans. J Bacteriol. 1981 Jul;147(1):62–68. doi: 10.1128/jb.147.1.62-68.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K. K., Newton A. Patterns of protein synthesis during development in Caulobacter crescentus. Dev Biol. 1977 Apr;56(2):417–425. doi: 10.1016/0012-1606(77)90281-0. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford I. P. Comparative studies on the regulation of tryptophan synthesis. CRC Crit Rev Biochem. 1980;8(2):175–189. doi: 10.3109/10409238009105468. [DOI] [PubMed] [Google Scholar]
- Crawford I. P., Gunsalus I. C. Inducibility of tryptophan synthetase in Pseudomonas putida. Proc Natl Acad Sci U S A. 1966 Aug;56(2):717–724. doi: 10.1073/pnas.56.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Amarasinghe A. B., Bender R. A. Ammonia assimilation and glutamate formation in Caulobacter crescentus. J Bacteriol. 1978 Jan;133(1):225–230. doi: 10.1128/jb.133.1.225-230.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Croft R. H. Transposon mutagenesis in Caulobacter crescentus. J Bacteriol. 1982 Feb;149(2):620–625. doi: 10.1128/jb.149.2.620-625.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Johnson R. C. Generalized Transduction in CAULOBACTER CRESCENTUS. Genetics. 1977 Nov;87(3):391–399. doi: 10.1093/genetics/87.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B. Transfer of drug resistance factors to the dimorphic bacterium Caulobacter crescentus. Genetics. 1979 Mar;91(3):371–380. doi: 10.1093/genetics/91.3.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus C., Gunsalus C. F., Chakrabarty A. M., Sikes S., Crawford I. P. Fine structure mapping of the tryptophan genes in Pseudomonas putida. Genetics. 1968 Nov;60(3):419–435. doi: 10.1093/genetics/60.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Ely B. Analysis of nonmotile mutants of the dimorphic bacterium Caulobacter crescentus. J Bacteriol. 1979 Jan;137(1):627–634. doi: 10.1128/jb.137.1.627-634.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Ely B. Isolation of spontaneously derived mutants of Caulobacter crescentus. Genetics. 1977 May;86(1):25–32. doi: 10.1093/genetics/86.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Milhausen M., Gill P. R., Parker G., Agabian N. Cloning of developmentally regulated flagellin genes from Caulobacter crescentus via immunoprecipitation of polyribosomes. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6847–6851. doi: 10.1073/pnas.79.22.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta N., Chen L. S., Newton A. Isolation and expression of cloned hook protein gene from Caulobacter crescentus. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4863–4867. doi: 10.1073/pnas.79.16.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POINDEXTER J. S. BIOLOGICAL PROPERTIES AND CLASSIFICATION OF THE CAULOBACTER GROUP. Bacteriol Rev. 1964 Sep;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawula R. V., Crawford I. P. Mapping of the tryptophan genes of Acinetobacter calcoaceticus by transformation. J Bacteriol. 1972 Nov;112(2):797–805. doi: 10.1128/jb.112.2.797-805.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Widera G., Gautier F., Lindenmaier W., Collins J. The expression of tetracycline resistance after insertion of foreign DNA fragments between the EcoRI and HindIII sites of the plasmid cloning vector pBR 322. Mol Gen Genet. 1978 Jul 25;163(3):301–305. doi: 10.1007/BF00271959. [DOI] [PubMed] [Google Scholar]
- Zalkin H., Murphy T. Utilization of ammonia for tryptophan synthesis. Biochem Biophys Res Commun. 1975 Dec 15;67(4):1370–1377. doi: 10.1016/0006-291x(75)90178-3. [DOI] [PubMed] [Google Scholar]