Abstract
In this study we have sought to characterize a committed myeloid progenitor cell line in an attempt to isolate general factors that may promote differentiation. We used cDNA representational difference analysis (RDA), which allows analysis of differential gene expression, to compare EML and EPRO cells. We have isolated nine differentially expressed cDNA fragments as confirmed by slot blot, Northern, and PCR analysis. Three of nine sequences appear to be novel whereas the identity of the remaining fragments suggested that the EPRO cell line is multipotent. Among the isolated sequences were eosinophilic, monocytic, and neutrophilic specific genes. Therefore, we tested the ability of EPRO cells to differentiate along multiple myeloid lineages and found that EPRO cells exhibited morphologic maturation into either monocyte/macrophages or neutrophils, but not eosinophils. Furthermore, when EPRO cells were exposed to ATRA, neutrophil specific genes were induced, whereas monocytic markers were induced by phorbol ester treatment. This study highlights the use of cDNA RDA in conjunction with the EML/EPRO cell line to isolate markers associated with macrophage and neutrophil differentiation and establishes the usefulness of this system in the search for factors involved in myeloid commitment.
Hematopoietic stem cells divide stochastically to give rise to daughter cells exhibiting proliferative capacity or committed progenitor cells with a more restricted range of differentiative capacity (1). As these committed cells proliferate, they gradually become restricted further and undergo terminal differentiation into a given cell type dependent on a variety of extrinsic and cell autonomous factors (1–3). Studies of leukemic cell lines provide evidence for a “nodal” decision point from which cells may differentiate along the monocyte or neutrophil lineage. Both the HL60 (4) and NB4 (5) cell lines were isolated from patients with acute promyelocytic leukemia. They appear morphologically as promyelocytes and mature into neutrophils upon exposure to all-trans retinoic acid (ATRA) (5–7), though they fail to express secondary granule protein transcripts, a marker of normal neutrophil maturation (8–10). Both of these cell lines also exhibit differentiation into macrophage-like cells upon induction with the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA) (4, 9). TPA induction of these cells causes adherence to plastic with pseudopodia formation, rapid loss of proliferative capacity, and induction of α-napthyl acetate esterase (nonspecific esterase). Interestingly, expression of neutrophil gelatinase also is rapidly up-regulated in response to TPA, whereas other secondary granule protein genes fail to be expressed (9).
The EML cell line was established by transducing mouse bone marrow with a retrovirus harboring a dominant-negative form of the retinoic acid receptor α (11). EML cells can be induced to undergo differentiation along the erythroid, lymphoid, and myeloid lineages, although maturation along the myeloid lineage requires the presence of ATRA at superphysiological concentrations. A more committed myeloid progenitor cell line can be derived from EML by inducing with ATRA and interleukin 3 (IL-3) in the presence of stem cell factor then culturing these cells in granulocyte/macrophage colony-stimulating factor (GM-CSF) alone (11). The resulting cell line, referred to as EPRO, is composed of promyelocytes that can undergo neutrophil maturation in response to ATRA. Furthermore, in contrast to HL60 and NB4, these cells also express secondary granule protein transcripts that are induced after exposure to ATRA.
Representational difference analysis (RDA) initially was established as a means to isolate differences in genomic DNA sequences and has been used to identify chromosomal aberrations as well as restriction fragment length polymorphism linkage markers (12, 13). The technique has since been modified to utilize cDNA as starting material, thereby allowing analysis of differential gene expression (14). In an attempt to isolate elements involved in myeloid lineage commitment as well as markers of myeloid differentiation, we employed the technique of RDA to compare EPRO cells with the parental EML cell line. In this study we describe the differentially expressed genes we have obtained as well as the characteristics of the EPRO cell line predicted by these sequences.
MATERIALS AND METHODS
Cell Lines.
EML, BHK/MKL, and HM-5 cell lines were kindly provided by Schickwann Tsai, Fred Hutchinson Cancer Research Center, Seattle, WA. EML cells were maintained in Iscove’s’ modified Dulbecco’s medium (IMDM) containing 20% heat-inactivated horse serum and supplemented with 10% conditioned medium from the BHK/MKL cell line containing rat stem cell factor. The EPRO cell line was derived as described previously (11) and maintained in IMDM with 20% horse serum and 10% HM-5 conditioned medium as a source of mouse GM-CSF. Single-cell clones were obtained by diluting to 3 cells/ml and plating in 96-well plates (Corning/Costar). Induction of neutrophil maturation in EPRO cells was accomplished by adding ATRA (Sigma) to normal growth medium at a final concentration of 10 μM. To induce cells toward the monocytic/macrophage lineage, TPA (Sigma) was added to a final concentration of 3.24 × 10−6 M in the presence of normal growth medium. For induction toward the eosinophilic lineage, recombinant murine IL-5 (R & D Systems) was added to cultures as described in the text.
Morphologic Analysis.
For analysis of uninduced and ATRA-induced EPRO cells, cytospin smears were made and stained with Wright–Giemsa. For analysis of monocytic differentiation, EPRO cells were placed in a Lab-Tek chamber slide (Nunc) just before TPA induction. After 24 h, the slides were stained with Wright–Giemsa. Eosinophil maturation was monitored by making cytospin smears at various time points and staining with Fast green/Neutral red (Sigma).
RNA Isolation and cDNA Synthesis.
Total RNA was isolated from EML and EPRO cells by using TRIzol reagent (Life Technologies, Gaithersburg, MD). Poly(A)+ RNA was purified further from total RNA by two passages over an oligo(dT) column (Life Technologies). Double-stranded cDNA was made from mRNA by using standard procedures (15).
cDNA RDA.
RDA was performed as described elsewhere (14). The driver cDNA was obtained from uninduced EML cells, whereas tester was from uninduced EPRO cells. A second round of RDA was performed after sequence identification of initial third-difference product (DP3) fragments. This was done by supplementing the driver with 50–100 ng of individually gel-purified DP3 fragments before hybridization.
Northern and Slot Blot Analysis.
Northern blot analysis was performed as described elsewhere (16, 17). Northern blots contained either 1 μg mRNA (confirmation of differential DP3 expression) or 10 μg total RNA (analysis of EPRO differentiation) per lane. For lactoferrin and β-actin, probes were labeled with digoxigenin-UTP by random prime and hybridized to blots overnight in DIG EasyHyb buffer (Boehringer Mannheim) at 65°C. All other probes were prepared and radiolabeled as described elsewhere (17). Northern blots hybridized to UTP-labeled probes were washed as usual, and signal detection was carried out according to the manufacturer’s protocols (Genius System, Boehringer Mannheim).
For slot blot analysis, miniprep plasmid DNA (1 μl) was incubated at 98°C for 5 min and then placed on ice, and SSC was added to a 10× final concentration. Samples were then immobilized on Hybond nylon membrane (Amersham) by using a vacuum slot blot apparatus (Schleicher & Schuell). The membranes then were soaked in 0.4 M NaOH, neutralized in 1.5 M NaCl/1 M Tris, pH 7.4, and baked for 2 h at 80°C. EML and EPRO cDNA (100 ng of each) were radiolabeled as above, and equal cpm of each probe were hybridized to duplicate slot blots in 50% formamide hybridization buffer at 42°C for 36 h. Both Northern and slot blots were washed as described elsewhere (17).
RESULTS
RDA Analysis of the EPRO Cell Line.
RDA was used to isolate cDNA fragments that were expressed specifically in EPRO cells. After an initial round of RDA, several EPRO-specific fragments were isolated. To confirm that these fragments were differentially expressed, we utilized three screens: (i) plasmids containing fragments from the DP3 were used to make duplicate slot blots, which subsequently were probed with radiolabeled cDNA from EML or EPRO cells; those fragments that were found to be differentially expressed were then confirmed by (ii) Northern analysis of EML and EPRO mRNA, or (iii) PCR of EML and EPRO cDNA.
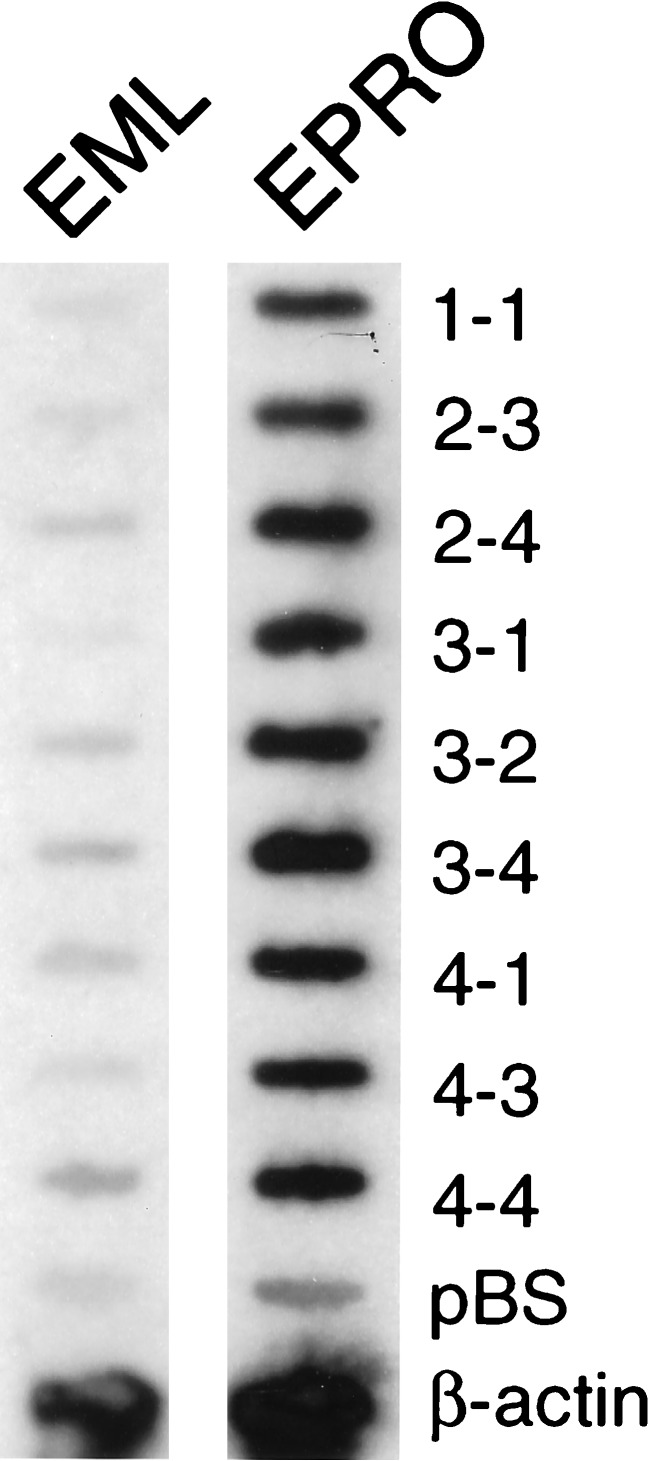
Fig. 1 shows a representative slot blot containing plasmids from the first round of RDA hybridized to radiolabeled EML or EPRO cDNA. Mouse β-actin and pBluescript were included on the blots as positive and negative controls, respectively. All of the initial plasmids isolated from the first round of RDA exhibit differential hybridization to the EPRO cDNA, i.e., they are present in the EPRO cDNA but not in the EML cDNA, whereas β-actin shows hybridization to both populations of cDNA (Fig. 1).
Figure 1.
Slot blot analysis of DP3 products. Plasmid DNA was denatured and transferred to duplicate nylon membranes. The blots were probed with equal cpm/ml of radiolabeled EML or EPRO cDNA. These are representative blots containing subcloned DP3 fragments from the first round of RDA. pBluescript and β-actin were included on the blot as negative and positive controls, respectively.
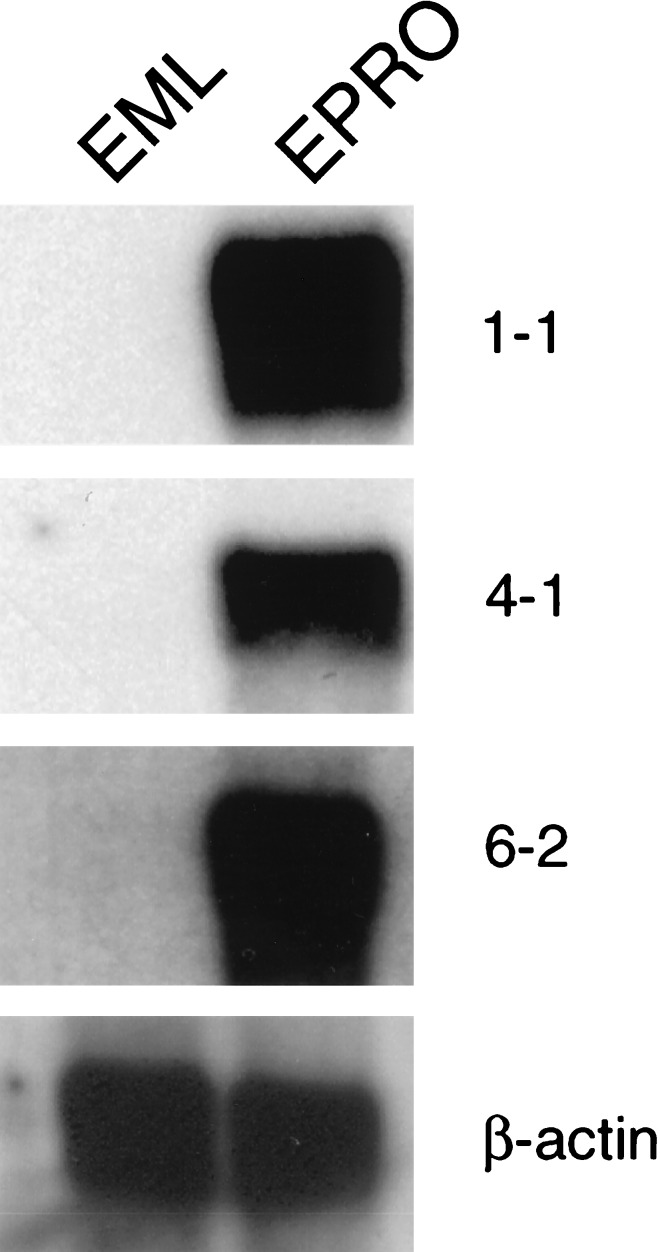
To further confirm that DP3 fragments were differentially expressed, they were excised from the plasmids, radiolabeled, and used as probes against Northern blots of mRNA from EML and EPRO cells. Fig. 2 shows the results of Northern analysis for EPRO 1–1, 4–1, and 6–2; all three are expressed in EPRO cells while completely absent from EML cells. To confirm that equal amounts of mRNA were present in each lane, the blots were probed subsequently with β-actin.
Figure 2.
Northern analysis of DP3 fragments. Subcloned DP3 fragments were used as probes against Northern blots of EML and EPRO RNA samples to confirm differential expression. Approximately 1 μg of mRNA was used for Northern analysis, and the blot was sequentially hybridized to DP3 fragments (1-1, 4-1, and 6-2 in this case). The blot was then hybridized with β-actin to confirm equal loading. The approximate size of each transcript is as follows: 1-1, a doublet of 1.3 and 1.6 kb; 4-1, 3.5 kb; 6-2, 2.5 kb.
After two rounds of RDA, a total of 30 fragments were isolated that exhibited differential expression by Northern blot and PCR analysis (data not shown). One clone was isolated that hybridized to both EML and EPRO mRNA.
Identification of RDA Products.
Fragments that were found to be differentially expressed were sequenced and compared with known GenBank sequences via a blast search to determine their identity. A total of nine genes, represented by 30 fragments, were isolated from two rounds of RDA. Table 1 lists the identity of the sequences isolated; interestingly, these genes are characteristic of eosinophil, neutrophil, and monocytic lineages. Those genes expressed in monocyte/macrophages include the galactose/N-acetylgalactoseamine-specific lectin isolated from mouse tumoricidal macrophages (18) and the putative cDNA for YM-1, a secreted glycoprotein isolated from activated peritoneal macrophages. C3, a component of the complement pathway (19), also was found to be expressed in EPRO cells, and its expression has been described in mouse macrophage-like cell lines (20). Neutrophil-specific genes include the IL-8 receptor (21), as well as a fragment bearing homology to human neutrophil collagenase. We subsequently have used this fragment (EPRO 6-2) to isolate a cDNA that is 73% identical to human neutrophil collagenase and represents the mouse homolog of this gene (22). The eosinophil lineage was found to be represented by eosinophil peroxidase (23). The three remaining gene fragments are novel; EPRO 7-3 and 8-5 do not align with any known sequence in GenBank or the Expressed Sequence Tag database (dbEST), whereas hypothetical translation of EPRO 10-4 reveals 53% amino acid identity to a putative murine inositol 5-phosphatase associated with the vibrator mutation (24) and likely represents a novel member of this family of proteins.
Table 1.
Sequence identity of EPRO-specific DP3 fragments
| Subclone | Description | Lineage | Genbank accession no. | Ref. |
|---|---|---|---|---|
| 1-1 | Putative cDNA for YM-1 | Mono/mac | M94584 | Unpublished data |
| 3-1 | Complement protein, C3 | Mono/mac | K02782 | (19, 20) |
| 4-1 | Eosinophil peroxidase, nucleotides 1412–1781 | Eosinophil | D78353 | (23) |
| 6-2 | Mouse neutrophil collagenase | Neutrophil | U96696 | (22) |
| 6-3 | Eosinophil peroxidase, nucleotides 1791–2336 | Eosinophil | — | — |
| 7-1 | Eosinophil peroxidase, nucleotides 454–935 | Eosinophil | — | — |
| 7-4 | No homology | ? | AF046876 | — |
| 8-5 | No homology | ? | AF046877 | — |
| 9-2 | Eosinophil peroxidase, nucleotides 1033–1389 | Eosinophil | — | — |
| 9-5 | Galactose/N-acetylgalactosamine-specific lectin | Mono/mac | S36676 | (18) |
| 10-2 | High-affinity IL-8 receptor | Neutrophil | D17630 | (21) |
| 10-4 | 53% amino acid identity to putative phosphoinositide 5-phosphatase type II | ? | AF046878 | (24) |
The EPRO Cell Line Is Bipotential.
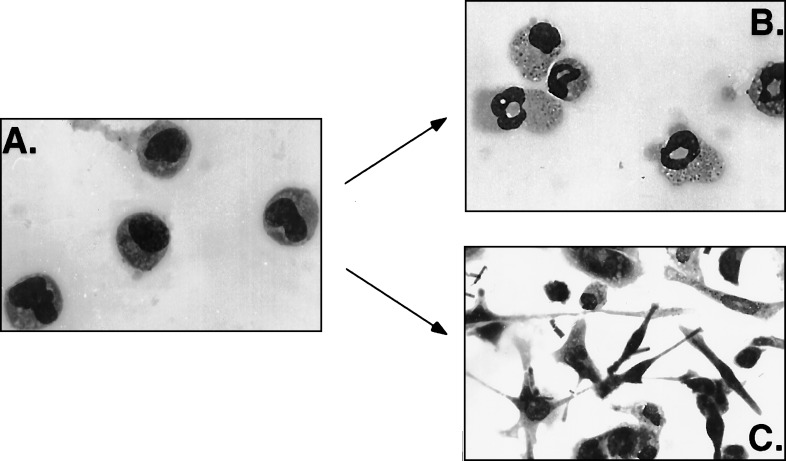
The identity of the genes isolated from the RDA screen provided evidence that the EPRO cell line may represent a multipotent cell line. To determine whether the EPRO cell line represented a multilineage progenitor, cells were induced with agents known to elicit monocytic, granulocytic, or eosinophilic differentiation in other systems. We confirmed the previously characterized ability of EPRO cells to undergo neutrophil maturation, and by 48 h of ATRA induction, cells begin to exhibit neutrophil morphology (Fig. 3B).
Figure 3.
Morphologic differentiation of EPRO cells. Uninduced EPRO cells (A) appear as promyelocytic cells that can undergo neutrophilic maturation in the presence of 10 μM ATRA (B; treated for 48 h) or monocytic differentiation in response to the phorbol ester, TPA (C, treated for 24 h).
To determine whether EPRO cells were also able to undergo monocytic differentiation, cells were induced with TPA for up to 48 h. After 24 h, an obvious morphological difference was noted between induced and uninduced cells (Fig. 3). The majority of uninduced EPRO cells grow in suspension (Fig. 3A) with some adherent cells; after 24 h in the presence of TPA, the majority of cells adhere to the plate and exhibit pseudopodia characteristic of macrophage-like cells (Fig. 3C). EPRO cells subsequently were cloned by limiting dilution, and three clones were analyzed. All three clonal cell lines exhibited neutrophil and monocytic maturation (data not shown).
Because EPRO cells also expressed eosinophil peroxidase and initial results indicated that they were bipotential, we sought to determine whether these cells also could give rise to eosinophils. EPRO cells were induced with purified recombinant IL-5 in the presence or absence of GM-CSF conditioned medium and with or without 10 μM ATRA. Cells were induced for up to 96 h, and cytospins were made at 24-h intervals. Cells were then stained by Wright–Giemsa or Fast green/Neutral red, which is specific for eosinophils. In the presence of IL-5 and GM-CSF without ATRA, the cells remained as promyelocytes throughout the induction. In the presence of IL-5, GM-CSF, and ATRA, the cells differentiated into neutrophils with no evidence of any eosinophils. Without GM-CSF and in the presence IL-5 with or without ATRA, the majority of cells died within 24 h with no evidence of eosinophilic differentiation. Despite the absence of eosinophil maturation in the presence of these cytokines, EPRO cells were also found to express eosinophil major basic protein (MBP) by reverse transcription–PCR analysis (data not shown).
Northern Analysis of ATRA- and TPA-Induced Cells.
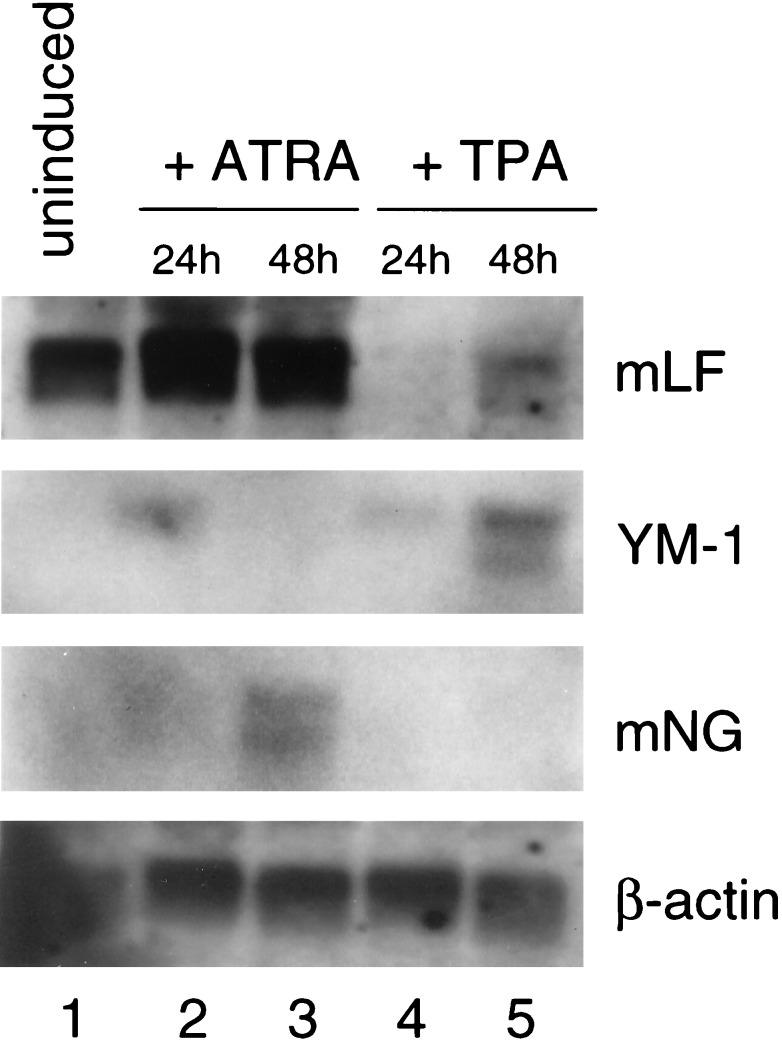
To further confirm the bipotential nature of EPRO cells, we performed Northern analysis on cells induced with ATRA or TPA by using neutrophil- and monocyte-specific markers. RNA was isolated for Northern analysis of uninduced EPRO cells and cells induced with either ATRA or TPA for 24 and 48 h. Fig. 4 shows results from both TPA and ATRA induction of bulk EPRO cells. The transcript for lactoferrin, a secondary granule protein, is up-regulated in response to ATRA in the closely related MPRO cell line (16) and in response to G-CSF in 32D Cl3 cells (17). To confirm neutrophil maturation of EPRO cells we analyzed lactoferrin expression by Northern analysis of ATRA and TPA induced EPRO cells. As seen in Fig. 1, lactoferrin (mLF) is expressed in uninduced cells and is up-regulated upon induction with ATRA (lanes 1–3). Upon induction with TPA, the transcript for mLF is potently down-regulated by 24 h (lane 4), yet increases again by 48 h (lane 5). In contrast, the transcript for YM-1, a macrophage marker, shows low-level expression in uninduced (also see Fig. 2, EPRO 1–1) and ATRA-induced cells (lanes 1–3) but is up-regulated at 24 and 48 h when induced with TPA (lanes 4 and 5). The discrepancy in YM-1 expression in uninduced cells is a result of the use of 1 μg of mRNA per lane in Fig. 2 compared with 10 μg total RNA in Fig. 4. Reverse transcription–PCR for c-fms (macrophage colony-stimulating factor receptor) shows an increase of expression from EML to EPRO and maintenance of expression during TPA induction, further confirming monocyte/macrophage potential (data not shown).
Figure 4.
Northern analysis of differential gene regulation in EPRO cells. Ten micrograms of total RNA was analyzed by Northern analysis by using probes for mouse lactoferrin (mLF), YM-1, mouse neutrophil gelatinase (mNG), and β-actin.
In NB4 and HL60 cells, induction with TPA is known to induce rapid up-regulation of human neutrophil gelatinase (25, 26), whereas in murine 32D Cl3 and MPRO cells, the corresponding mouse neutrophil gelatinase is up-regulated late in granulocytic differentiation (16, 17). We determined whether neutrophil gelatinase was induced along both lineages in the EPRO cell system by reprobing the Northern blot with a fragment of mouse neutrophil gelatinase (mNG). Fig. 4 shows that, as expected, neutrophil gelatinase is induced beginning at 48 h after induction with ATRA (lane 3), but mNG is not up-regulated in response to TPA (lanes 4 and 5).
DISCUSSION
In this study we describe the characterization of a myeloid progenitor cell line by use of cDNA RDA. Our initial goal was to isolate factors that were involved in myeloid determination. To this end we utilized the EML cell line, a previously characterized, multipotent cell line from which a committed progenitor cell line (EPRO) can be obtained.
We have found that EPRO cells express genes that are characteristic of multiple myeloid lineages. In agreement with previous findings that EPRO cells are neutrophilic, our RDA screen revealed specific expression of neutrophil collagenase and the IL-8 receptor. We subsequently have isolated and characterized a full-length clone of mouse neutrophil collagenase and have shown that it is up-regulated upon ATRA-induced neutrophil maturation in the closely related MPRO cell line (16, 22, 27).
Because our study also revealed genes characteristic of the monocyte/macrophage lineage, we sought to determine whether EPRO cells possessed multilineage potential. When induced with TPA, EPRO cells exhibit morphologic maturation to macrophages, including adherence and pseudopodia formation. Furthermore, a macrophage marker isolated from the RDA screen, YM-1, is specifically up-regulated during monocytic, but not neutrophilic, maturation. In support of our observation of monocytic potential, we found that both EML and EPRO cells express c-fms, although it has been shown that wild-type EML cells are not capable of responding to M-CSF and overexpression of c-fms in EML cells results in proliferation but not monocyte/macrophage commitment in response to M-CSF (28).
In contrast to monocyte markers, lactoferrin expression is potently down-regulated upon TPA induction, although expression is apparent at 48 h after TPA induction. The appearance of lactoferrin transcript at this time point may represent proliferating cells that were not responsive to TPA and retain this characteristic of uninduced EPRO cells. The high level of lactoferrin expression in uninduced cells likely is a result of the high concentration of horse serum in the growth medium, which we have found responsible for increased differentiation and lactoferrin expression in the related MPRO cell line (16).
Surprisingly, the level of neutrophil gelatinase, which has been shown to be potently up-regulated by TPA in NB4 cells (9), was not affected by TPA in EPRO cells. The reason for this observation is unknown but may be because of interference of some aspects of TPA signaling by the dominant-negative RARα expressed in these cells.
Although eosinophil peroxidase was found to be expressed, implying the presence of an early eosinophilic precursor, we were unable to observe terminal eosinophilic maturation in the presence of IL-5. There may be several explanations as to why we did not observe eosinophil maturation. Although GM-CSF and IL-5 have been shown to synergistically interact to induce eosinophil colony formation in mouse bone marrow (29), we did not observe eosinophil maturation of EPRO cells in response to IL-5. It has been shown that IL-3 also can act synergistically with GM-CSF and IL-5 and may act as a more potent inducer of eosinophil progenitor formation (30). It may be that IL-3 is required in conjunction with both IL-5 and GM-CSF to induce eosinophil formation in EPRO cells. Another possibility is that eosinophil peroxidase expression is artifactual and these cells possess no eosinophilic potential, though expression of eosinophil MBP in EPRO cells weakens this argument. Alternatively, the dominant-negative RARα may interfere with terminal eosinophil maturation, though in a retinoid-independent fashion, because retinoic acid has been shown to antagonize eosinophilic differentiation (31).
The bilineage potential of leukemic cell lines, such as HL60 and NB4, has been demonstrated in the past (4, 9). Furthermore, a recently described acute promyelocytic leukemia-derived cell line was shown to possess both neutrophilic and eosinophilic potential (32). However, in HL60 and NB4 cells, maturation is associated with abnormalities in late gene expression, which makes the relationship of these results to normal myeloid cell maturation uncertain (8, 9). In these studies we have shown that a factor-dependent, nontransformed cell line shows similar bilineage potential. Although this line too is abnormal, in that it expresses the dominant-negative RARα, the evidence for normal expression of late-stage- and cell-type specific genes (11, 16, 22) suggests that it better mimics the normal myeloid differentiation program. These studies therefore lend support to a model in which there is a common promyelocytic precursor to monocytes, neutrophils, and possibly eosinophils. In addition, this work highlights the usefulness of the EML/EPRO cell system in characterizing the progression of commitment during hematopoiesis. Because we were able to isolate specific markers of late stages in myeloid differentiation, it can be inferred that the factors responsible for the expression of these genes also are present. Thus, in conjunction with further rounds of cDNA RDA this system may provide a powerful tool to dissect the molecular mechanisms of myeloid differentiation.
Acknowledgments
We thank Schickwann Tsai for providing EML cells and members of the Berliner Lab for helpful discussion. N.B. is supported by National Institutes of Health Grants DK-48053 and HD33184 and by a scholar grant from the Leukemia Society of America.
ABBREVIATIONS
- ATRA
all-trans retinoic acid
- RDA
representational difference analysis
- TPA
12-O-tetradecanoylphorbol-13-acetate
- IL
interleukin
- GM-CSF
granulocyte/macrophage colony-stimulating factor
- DP3
third-difference product
Footnotes
References
- 1.Ogawa M. Blood. 1993;81:2844–2853. [PubMed] [Google Scholar]
- 2.Kehrl J H. Stem Cells. 1995;13:223–241. doi: 10.1002/stem.5530130304. [DOI] [PubMed] [Google Scholar]
- 3.Metcalf D. Blood. 1993;82:3515–3523. [PubMed] [Google Scholar]
- 4.Collins S J. Blood. 1987;70:1233–1244. [PubMed] [Google Scholar]
- 5.Lanotte M, Martin-Thouvenin V, Najman S, Balerini P, Valensi F, Berger R. Blood. 1991;5:1080–1086. [PubMed] [Google Scholar]
- 6.Collins S J, Gallo R C, Gallagher R E. Nature (London) 1977;270:347–349. doi: 10.1038/270347a0. [DOI] [PubMed] [Google Scholar]
- 7.Collins S J, Ruscetti F W, Gallagher R E, Gallo R C. Proc Natl Acad Sci USA. 1978;75:2458–2462. doi: 10.1073/pnas.75.5.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnston J J, Rintels P, Chung J, Sather J, Benz E J, Jr, Berliner N. Blood. 1992;79:2998–3006. [PubMed] [Google Scholar]
- 9.Khanna-Gupta A, Kolibaba K, Zibello T A, Berliner N. Blood. 1994;84:294–302. [PubMed] [Google Scholar]
- 10.Khanna-Gupta A, Zibello T, Berliner N. Curr Top Microbiol Immunol. 1996;211:165–171. doi: 10.1007/978-3-642-85232-9_17. [DOI] [PubMed] [Google Scholar]
- 11.Tsai S, Bartelmez S, Sitnicka E, Collins S. Genes Dev. 1994;8:2831–2841. doi: 10.1101/gad.8.23.2831. [DOI] [PubMed] [Google Scholar]
- 12.Lisitsyn N, Lisitsyn N, Wigler M. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 13.Lisitsyn N A, Segre J A, Kusumi K, Lisitsyn N M, Nadeau J H, Frankel W N, Wigler M H, Lander E S. Nat Genet. 1994;6:57–63. doi: 10.1038/ng0194-57. [DOI] [PubMed] [Google Scholar]
- 14.Hubank M, Schatz D G. Nucleic Acids Res. 1994;22:5640–5648. doi: 10.1093/nar/22.25.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Greene & Wiley; 1987. [Google Scholar]
- 16.Lawson, N. D., Krause, D. S. & Berliner, N. (1998) Exp. Hematol., in press. [PubMed]
- 17.Graubert T, Johnston J, Berliner N. Blood. 1993;82:3192–3197. [PubMed] [Google Scholar]
- 18.Sato M, Kawakami K, Osawa T, Toyoshima S. J Biochem. 1992;111:331–336. doi: 10.1093/oxfordjournals.jbchem.a123758. [DOI] [PubMed] [Google Scholar]
- 19.Fey G H, Lundwall A, Wetsel R A, Tack B F, de Bruijn M H, Domdey H. Philos Trans R Soc London B Biol Sci. 1984;306:333–344. doi: 10.1098/rstb.1984.0094. [DOI] [PubMed] [Google Scholar]
- 20.Fey G, Domdey H, Wiebauer K, Whitehead A S, Odink K. Springer Semin Immunopathol. 1983;6:119–147. doi: 10.1007/BF00205869. [DOI] [PubMed] [Google Scholar]
- 21.Harada A, Kuno K, Nomura H, Mukaida N, Murakami S, Matsushima K. Gene. 1994;142:297–300. doi: 10.1016/0378-1119(94)90278-x. [DOI] [PubMed] [Google Scholar]
- 22.Lawson N D, Khanna-Gupta A, Berliner N. Blood. 1998;91:2517–2524. [PubMed] [Google Scholar]
- 23.Horton M A, Larson K A, Lee J J, Lee N A. J Leukocyte Biol. 1996;60:285–294. doi: 10.1002/jlb.60.2.285. [DOI] [PubMed] [Google Scholar]
- 24.Hamilton B A, Smith D J, Mueller K L, Kerrebrock A W, Bronson R T, van Berkel V, Daly M J, Kruglyak L, Reeve M P, Nemhauser J L, et al. Neuron. 1997;18:711–722. doi: 10.1016/s0896-6273(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 25.Kimmel C B, Ballard W W, Kimmel S R, Ullmann B, Schilling T F. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 26.Devarajan P, Johnston J J, Ginsberg S S, Van Wart H E, Berliner N. J Biol Chem. 1992;267:25228–25232. [PubMed] [Google Scholar]
- 27.Tsai S, Collins S J. Proc Natl Acad Sci USA. 1993;90:7153–7157. doi: 10.1073/pnas.90.15.7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai S. Cancer Chemother Pharmacol. 1996;38:S58–S63. doi: 10.1007/s002800051040. [DOI] [PubMed] [Google Scholar]
- 29.Takamoto M, Sugane K. Immunol Lett. 1995;45:43–46. doi: 10.1016/0165-2478(94)00223-e. [DOI] [PubMed] [Google Scholar]
- 30.Warren D J, Moore M A. J Immunol. 1988;140:94–99. [PubMed] [Google Scholar]
- 31.Paul C C, Mahrer S, Tolbert M, Elbert B L, Wong I, Ackerman S J, Baumann M A. Blood. 1995;86:3737–3744. [PubMed] [Google Scholar]
- 32.Kishi K, Toba K, Azegami T, Tsukada N, Uesugi Y, Masuko M, Niwano H, Hashimoto S, Sakaue M, Furukawa T, et al. Exp Hematol. 1998;26:135–142. [PubMed] [Google Scholar]