Abstract
From Toki-shakuyaku-san, an herbal formulation for “cleansing stagnated blood,” a key gene regulatory compound was purified and identified through a screening based on DNA microarray and real-time PCR.
An Oriental herbal formulation, Toki-shakuyaku-san (TSS), has long been used for the treatments of various diseases related to circulation problems, including gynecological and obstetric disorders.1,2 The formulation is made from six different herbs: Hoelin (Fu ling), Cnidium Rhizome (Chuan xiong), Angelica Sinensis (Dang gui), Peony Root (Shao yao), Atractylodes Rhizome (Bai zhu) and Alisma Rhizome (Ze xie). In the parlance of the Oriental medicine, TSS is a formulation to “cleanse stagnated blood,” thereby nourishing the “yang” of blood. In fact, this somewhat nebulous claim is scientifically supported by the beneficial effects of TSS. The formulation exhibits therapeutic effects for various disorders associated with poor blood circulation, such as hot flashes,2 and preeclampsia.3 Clearly, TSS contains compounds that can improve blood circulation, although their identities have not been fully clarified.
Here, we conducted a screening of TSS to uncover previously overlooked bioactive compounds. To this end, this study employed a protocol called “genomic screening,” which was recently established in our group.4 In genomic screening, biologically active compounds are detected and purified based on their ability to regulate cellular messenger RNA (mRNA) expression. DNA microarray is used first to identify the most prominent cellular transcriptional responses to a mixture of compounds, such as herbal formulations. Real-time PCR of the identified genes are then used to guide the purification of active compounds. Although DNA microarray has been employed for the analyses of natural products,5 what sets this study apart from others is the employment of DNA microarray for the purpose of purifying biologically active compounds. The current screening uncovered a polyacetylene compound capable of regulating genes associated with blood coagulation in endothelial cells.
As the first step of genomic screening, we determined the genes most distinctly regulated by TSS. Human umbilical vein endothelial cells (HUVEC) were treated with 100 μg/ml of TSS for 4 hours. Expression profiling with Affymetrix GeneChip™ Human Genome U133 Plus 2.0 array revealed that, out of over 47,000 transcripts on the array, 92 genes were differentially regulated by TSS. Table 1 summarizes the most distinctly regulated genes (see also Supplementary data). Among these genes was SerpinB2, which is also known as plasminogen activator inhibitor type 2 (PAI-2). Since SerpinB2 is a well-characterized gene in terms of its roles in blood coagulation and circulation disorders,6,7 we decided to pursue the TSS component that induces this gene.
Table 1.
Endothelial genes most distinctly regulated by TSS (100 μg/ml, 4 hours)
| UniGene ID | Gene | Log ratioa | Fold Change | Changeb | Change p-valuec |
|---|---|---|---|---|---|
| Hs.12813 | TCDD-inducible poly(ADP-ribose) polymerase | 0.7 | 1.6 | I | 0.00002 |
| Hs.196384 | Prostaglandin-endoperoxide synthase 2 (COX2) | 1.0 | 2.0 | I | 0.00002 |
| Hs.594481 | Serpin peptidase inhibitor, clade B, member 2 (SerpinB2) | 1.0 | 2.0 | I | 0.00002 |
| Hs.433791 | Transmembrane protein 46 | 0.9 | 1.9 | I | 0.00002 |
| Hs.326035 | Early growth response 1 | 0.8 | 1.7 | I | 0.00002 |
| Hs.651231 | Heparan sulfate proteoglycan 2 (perlecan) | −0.7 | −1.6 | D | 0.99904 |
| Hs.594486 | Unknown | −1.9 | −3.7 | D | 0.99944 |
| Hs.29055 | Guanine nucleotide binding protein-like 3 | −0.8 | −1.7 | D | 0.99955 |
| Hs.557980 | Unknown | −0.7 | −1.6 | D | 0.99996 |
| Hs.355753 | 1-acylglycerol-3-phosphate O-acyltransferase 6 | −1.2 | −2.3 | D | 0.99998 |
Log ratio: Log2 (signal in TSS-treated HUVEC/signal in control HUVEC)
Change: Direction of change determined by Affymetrix Microarray Suite Version 5.0 (MAS5) Software
Change p-value: Log p-values are calculated using the Wilcoxon signed rank test and Tukey Biweight on MAS5. MAS5 calls a gene “decreased (D)” when its p-value between control and drug-treated groups is between 1 and 0.997, whereas a gene is considered “increased (I)” when its p-value is between 0 and 0.003. Shown on Table 1 are the top five up and down regulated genes based on the Change p-values.
The real-time PCR assay of SerpinB2 was used to guide the purification of active compound(s). Fractionation of TSS was carried out as outlined in Figure 1a. Individual fractions were submitted to the real-time PCR assay to examine their ability to induce SerpinB2. Activity was observed in the fractions indicated by the red arrows in Figure 1a. After several steps of fractionation, the active compound was isolated by reversed phase HPLC. The purified compound gave a single LC/MS peak (Fig. 1b). At the concentration of 5 μg/ml the purified compound caused approximately 2-fold induction (log2RQ ~1) of SerpinB2 in HUVEC (Fig. 1c).
Figure 1.
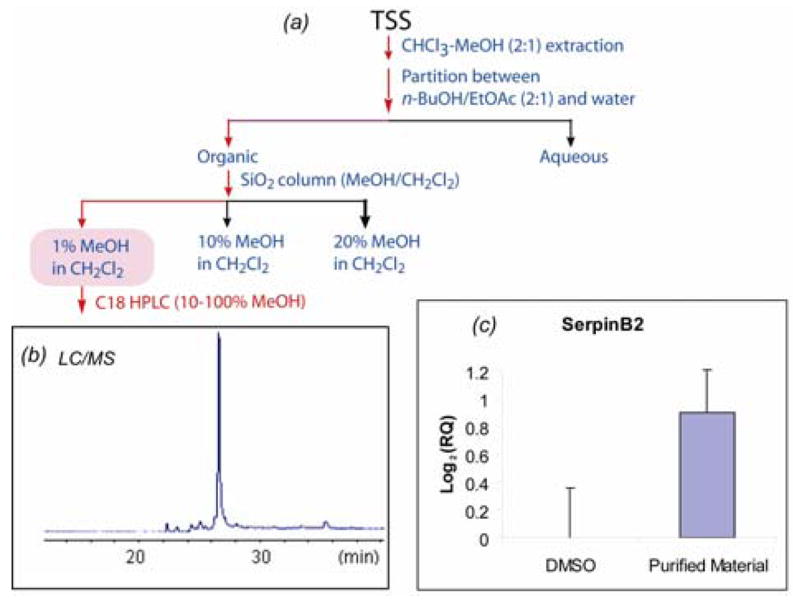
(a) Real-time RT-PCR (SERPINB2) guided purification of a gene regulatory compound from TSS. (b) LC/MS positive ion profile of the purified active compound. (c) SerpinB2 Real-time PCR assay of the active compound (5 μg/ml). GAPDH was used as the endogenous control. Log2(RQ) corresponds to the Log Ratio value in the expression profile (Table 1). RQ stands for relative quantitation (i.e., the fold-change ratio) in real-time PCR analysis.
The identity of the active compound was determined by spectroscopic analyses using high resolution mass spectrometry (HRMS) and NMR. Briefly, the molecular formula of the active compound was determined to be C18H22O4 based on HRMS. A partial structure was deduced by 1H, HSQC, and COSY (Figs 2a and 2b). SciFinder search identified one compound with a matching molecular formula (C18H22O4) and the partial structure (Fig 2b). This compound, (6E,12E)-tetradecadiene-8,10-diyne-1,3-diol diacetate (TDEYA), is a known component in Atractylodes Rhizome (Fig 2c).8 The 13C-NMR spectrum of the purified compound confirmed its identity to be indeed TDEYA. Little is known about the biological properties of TDEYA except for a modest inhibitory activity against xanthine oxidase (IC50 = 1.0 × 10−4 M).9
Figure 2.
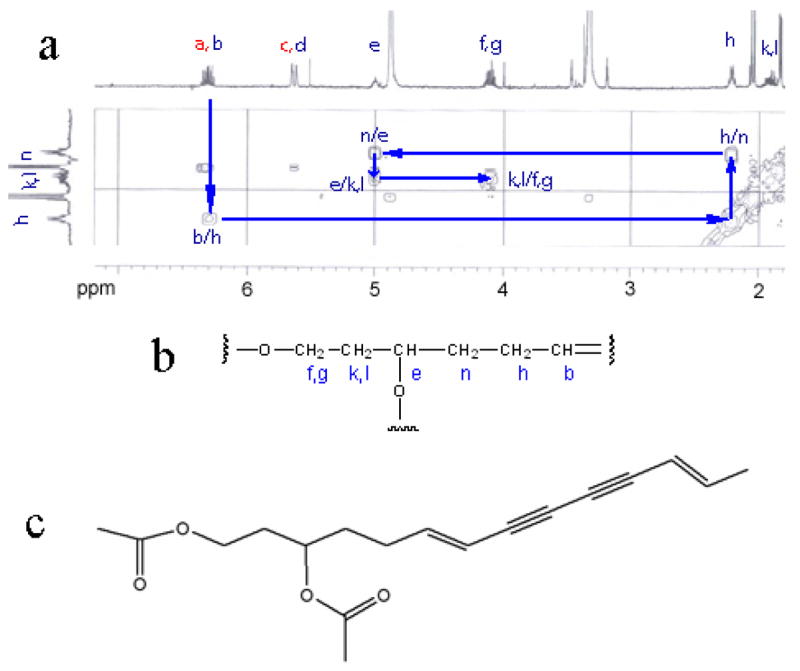
(a) COSY correlations of the purified active compound. (b) The partial fragment structure deduced based on 1H NMR, HSQC and COSY spectra. (c) The identified structure of the active compound.
The identification of TDEYA as the regulator of SerpinB2 made us wonder whether TDEYA regulates additional genes, especially those observed in the original expression profile of TSS (Table 1). To address this issue, we conducted mRNA expression profiling of TDEYA. Table 2 summarizes the most distinctly regulated genes by TDEYA in HUVEC. It turned out that many genes overlapped between the TSS and TDEYA profiles (e.g., the genes highlighted in bold in Table 2; also see Supplementary data), indicating that TDEYA is a key gene regulatory compound in TSS. Among the overlapped genes, regulation of perlecan, COX2 and TI-PARP by TDEYA was further validated by real-time PCR (see Supplementary data).
Table 2.
Endothelial genes most distinctly regulated by the purified active compound (TDEYA; 5 μg/ml, 4 hours)
| UniGene ID | Gene | Log ratioa | Fold Change | Changeb | Change p-valuec |
|---|---|---|---|---|---|
| Hs.12813 | TCDD-inducible poly(ADP-ribose) polymerase (TI-PARP) | 1.2 | 2.3 | I | 0.00002 |
| Hs.196384 | Prostaglandin-endoperoxide synthase 2 (COX2) | 1.0 | 2.0 | I | 0.00002 |
| Hs.594481 | Serpin peptidase inhibitor, clade B, member 2 (SerpinB2) | 1.0 | 2.0 | I | 0.00002 |
| Hs.154654 | Cytochrome P450, family 1, subfamily B, polypeptide 1 | 2.7 | 6.5 | I | 0.00002 |
| Hs.390594 | Solute carrier family 7, member 11 | 1.2 | 2.3 | I | 0.00002 |
| Hs.591945 | Zinc finger and BTB domain containing 16 | −1.0 | −2.0 | D | 0.99992 |
| Hs.124649 | ATP-binding cassette, sub-family G, member 1 | −0.9 | −1.9 | D | 0.99995 |
| Hs.651231 | Heparan sulfate proteoglycan 2 (perlecan) | −0.8 | −1.7 | D | 0.99996 |
| Hs.170355 | Mesenchyme homeobox 2 | −1.1 | −2.1 | D | 0.99997 |
| Hs.468972 | AT rich interactive domain 1A (SWI-like) | −0.9 | −1.9 | D | 0.99998 |
Since TSS is a putative “blood cleansing” formulation, we looked into literature to see if the overlapped genes have any links to blood coagulation. It turned out that three genes, i.e., SerpinB2, perlecan and COX2, are indeed associated with blood coagulation. SerpinB2 is sometimes considered as a “coagulation factor” because of its ability to inhibit plasminogen activator. However, insufficient SerpinB2 is actually linked to disorders associated with poor blood circulation; for example, low plasma level of SerpinB2 in pregnant women is a prognostic marker for decreased placental function and intrauterine growth retardation.7,10 Perlecan is an extracellular matrix protein involved in various vascular processes, such as angiogenesis and wound healing.11 A recent study found that perlecan is down-regulated in endothelial cells treated by antithrombin, which inactivates several enzymes of the coagulation system.12 COX-2 is a pro-inflammatory gene involved in the biosynthesis of prostaglandins. However, the anti-thrombotic function of COX-2 was revealed recently through the increased risk of thrombotic cardiovascular events among the patients treated with COX-2 inhibitors.13 Taken together, the links of these genes to blood coagulation suggest that the putative “blood cleansing” activity of TSS is mediated, in part, at the transcriptional level.
Among the known constituents in TSS, paeoniflorin and ferulic acid have been reported to slow thrombosis in animal models.14 Behind these compounds, however, TDEYA has been quietly regulating the genes related to blood coagulation. The current finding warrants further studies to define the biological properties of TDEYA, which, in turn, can provide novel insights into the roles of the observed genes in thrombosis.
In conclusion, genomic screening identified TDEYA, as a key gene regulatory compound in TSS. TDEYA regulates the expression of genes associated with blood coagulation. Further studies are needed to determine the therapeutic utilities of TDEYA.
Supplementary Material
Supplementary data
Supplementary data associated with this article can be found in the online version.
Acknowledgments
This study was supported in part by NIH GM60654. RR-03037 from NCRR/NIH, which partially supports the research infrastructure at Hunter College, is also acknowledged. LC/MS instrumentation was funded by NCRR/NIH RR022649-01. We thank Mr. Tal H. Hasson for his technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.Koyama T. J Jpn Menopause Soc. 1993;1:75. [Google Scholar]; Kano T, Ito C, Kasamatsu HYM. Jpn J Fertil Steril. 1991;36:612. [Google Scholar]; Kotani N, Oyama T, Sakai I, Hashimoto H, Muraoka M, Ogawa Y, Matsuki A. Am J Chin Med. 1997;25:205. doi: 10.1142/S0192415X9700024X. [DOI] [PubMed] [Google Scholar]
- 2.Tanaka T. Clin Exp Obstet Gynecol. 2001;28:20. [PubMed] [Google Scholar]
- 3.Takei H, Nakai Y, Hattori N, Yamamoto M, Kurauchi K, Sasaki H, Aburada M. Phytomedicine. 2004;11:43. doi: 10.1078/0944-7113-00332. [DOI] [PubMed] [Google Scholar]; Takei H, Yamamoto M, Kase Y, Takeda S. J Pharmacol Sci. 2005;98:255. doi: 10.1254/jphs.fpj04043x. [DOI] [PubMed] [Google Scholar]
- 4.Kawamura A, Brekman A, Grigoryev Y, Hasson TH, Takaoka A, Wolfe S, Soll CE. Bioorg Med Chem Lett. 2006;16:2846. doi: 10.1016/j.bmcl.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 5.Zhang YB, Wang J, Wang ZT, But PP, Shaw PC. Planta Med. 2003;69:1172. doi: 10.1055/s-2003-818015. [DOI] [PubMed] [Google Scholar]; Iizuka N, Oka M, Yamamoto K, Tangoku A, Miyamoto K, Miyamoto T, Uchimura S, Hamamoto Y, Okita K. Int J Cancer. 2003;107:666. doi: 10.1002/ijc.11452. [DOI] [PubMed] [Google Scholar]; Coldren CD, Hashim P, Ali JM, Oh SK, Sinskey AJ, Rha C. Planta Med. 2003;69:725. doi: 10.1055/s-2003-42791. [DOI] [PubMed] [Google Scholar]; Watanabe CM, Supekova L, Schultz PG. Chem Biol. 2002;9:245. doi: 10.1016/s1074-5521(02)00103-5. [DOI] [PubMed] [Google Scholar]
- 6.Kruithof EK, Baker MS, Bunn CL. Blood. 1995;86:4007. [PubMed] [Google Scholar]; Medcalf RL, Stasinopoulos SJ. FEBS J. 2005;272:4858. doi: 10.1111/j.1742-4658.2005.04879.x. [DOI] [PubMed] [Google Scholar]
- 7.Astedt B, Lindoff C, Lecander I. Semin Thromb Hemost. 1998;24:431. doi: 10.1055/s-2007-996035. [DOI] [PubMed] [Google Scholar]
- 8.Kano Y, Komatsu K, Saito K, Bando H, Sakurai T. Chem Pharm Bull. 1989;37:193. [Google Scholar]
- 9.Sakurai T, Yamada H, Saito K, Kano Y. Biol Pharm Bull. 1993;16:142. doi: 10.1248/bpb.16.142. [DOI] [PubMed] [Google Scholar]; Sakurai T, Sugawara H, Saito K, Kano Y. Biol Pharm Bull. 1994;17:1364. doi: 10.1248/bpb.17.1364. [DOI] [PubMed] [Google Scholar]
- 10.Estelles A, Gilabert J, Espana F, Aznar J, Galbis M. Am J Obstet Gynecol. 1991;165:138. doi: 10.1016/0002-9378(91)90242-j. [DOI] [PubMed] [Google Scholar]; Gilabert J, Estelles A, Ayuso MJ, Espana F, Chirivella M, Grancha S, Mico JM, Aznar J. Gynecol Obstet Invest. 1994;38:157. doi: 10.1159/000292470. [DOI] [PubMed] [Google Scholar]; Grancha S, Estelles A, Gilabert J, Chirivella M, Espana F, Aznar J. Thromb Haemost. 1996;76:761. [PubMed] [Google Scholar]
- 11.Knox SM, Whitelock JM. Cell Mol Life Sci. 2006;63:2435. doi: 10.1007/s00018-006-6162-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W, Chuang YJ, Swanson R, Li J, Seo K, Leung L, Lau LF, Olson ST. Blood. 2004;103:1185. doi: 10.1182/blood-2003-08-2920. [DOI] [PubMed] [Google Scholar]
- 13.Iezzi A, Ferri C, Mezzetti A, Cipollone F. Curr Pharm Des. 2007;13:1715. doi: 10.2174/138161207780831293. [DOI] [PubMed] [Google Scholar]
- 14.Ye J, Duan H, Yang X, Yan W, Zheng X. Planta Med. 2001;67:766. doi: 10.1055/s-2001-18364. [DOI] [PubMed] [Google Scholar]; Wang BH, Ou-Yang JP. Cardiovasc Drug Rev. 2005;23:161. doi: 10.1111/j.1527-3466.2005.tb00163.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data associated with this article can be found in the online version.


