Abstract
X chromosome inactivation involves a random choice to silence either X chromosome early in mammalian female development. Once silenced the inactive X is stably inherited through subsequent somatic cell divisions, and thus, females are generally mosaics, having a mixture of cells with one or the other parental X active. While in most females the number of cells with either X being active is roughly equal, skewing of X chromosome inactivation is observed in a percentage of women. In this issue of the JCI, Bolduc and colleagues address whether skewing of X chromosome inactivation in humans is influenced by an X-linked locus that can alter this initial random inactivation (see the related article beginning on page 333). Their data indicate that most of the skewing observed in humans results from secondary events rather than being due to an inherited tendency to inactivate a particular X chromosome.
What is skewed X chromosome inactivation?
X chromosome inactivation (XCI) — a process originally hypothesized by Lyon in 1961 (1) and by which one of the two copies of the X chromosome present in females is inactivated — achieves dosage equivalency for X-linked genes between XY males and XX females. A critical tenet of this hypothesis was that the initial choice of which X (maternal or paternal) to inactivate was random but then this choice was stably inherited. An individual cell’s decision to inactivate either the paternal or maternal X is made early in development, at approximately the time of implantation (2). A deviation from equal (50%) inactivation of each parental allele is known as skewing, with common criteria for “skewed” inactivation being arbitrarily defined as the observation of inactivation of the same allele in 75% or 80% of cells (and very skewed inactivation resulting in 90% or 95% of cells with the same allele inactive).
A solid understanding of the causes of skewed XCI is needed because skewing is often used as a tool in the clinical setting (see The clinical relevance of skewed XCI). As shown in Figure 1A, some individuals will undergo skewed XCI by chance, as there is a limited pool of cells at the time of inactivation and only a limited precursor population that contributes to any tissue. Selection can also lead to a secondary (or acquired) skewed inactivation. In addition, there may be variants controlling the initial choice of which X to inactivate, resulting in primary nonrandom inactivation. For example, in mice there is a locus known as the X chromosome controlling element (Xce) that differs between strains and results in primary skewing of inactivation during animal crosses (3). The murine Xce is located downstream of X (inactive)–specific transcript (Xist) (4), a gene that codes for a non–protein coding RNA that localizes to the inactive X chromosome and has been shown to be sufficient for XCI (reviewed in ref. 5). Whether there is a similar locus involved in XCI skewing in humans remains an open question. In this issue of the JCI, the study by Bolduc et al. addresses the relative contribution to XCI skewing of primary and secondary influences by investigating skewing in a population of over 500 pairs of healthy female mothers and their neonates (6).
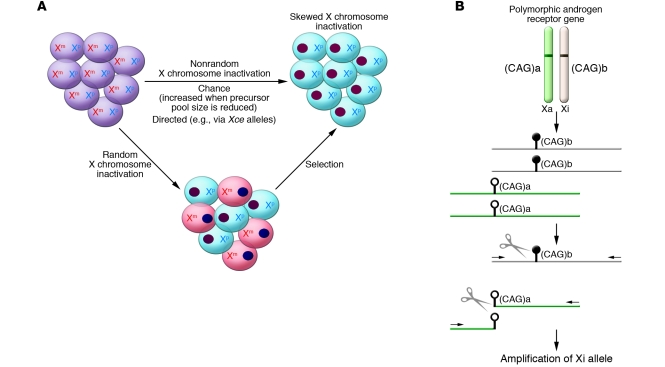
Figure 1. Skewed XCI in females.
(A) Skewed XCI can result from either a primary nonrandom inactivation or a secondary, acquired nonrandom inactivation. There are two active X chromosomes in the female embryo, one from each parent (maternal and paternal; Xm and Xp, respectively). Upon inactivation, one X forms the heterochromatic inactive X depicted here as a darkly staining oval. The cells are shaded to reflect the different origins of their remaining active X. Subsequent stochastic or selective growth of one cell population leads to skewing of inactivation. However, if the initial inactivation was influenced by an allele (analogous to the murine Xce allele) then inactivation could be directed primarily towards one allele. A reduction in the embryonic precursor pool size at the time of random choice would make skewed XCI more likely. (B) The common way to examine XCI skewing makes use of a highly polymorphic CAG repeat (shown here as having allele sizes a and b) in the androgen receptor gene and differential methylation (methylation shown as black lollipop while the unmethylated allele has open lollipops) of the active (Xa) and inactive (Xi) X. DNA is extracted from the cells to be tested and then digested with a methylation-sensitive restriction enzyme (shown as scissors). The methylated (Xi) DNA will remain intact for subsequent PCR amplification using primers (shown as arrows) flanking the cut site. The ratio of the alleles (a and b, distinguished by PCR product length) in the undigested compared with the digested DNA reflects the XCI skewing.
Assays to examine XCI skewing require the presence of a polymorphism to distinguish the two Xs and a means of determining which X is active. While examining RNA or protein is a more direct measure of X chromosome activity, DNA methylation of the inactive X is often used as a surrogate since DNA is more easily extracted, stored, and analyzed. Furthermore, coding region polymorphisms are generally less informative and can show allelic imbalances in expression levels. An assay monitoring DNA methylation of the highly polymorphic CAG trinucleotide polymorphism at the 5ι end of the gene encoding the androgen receptor, as used in the Bolduc study (6), has become the standard for measuring XCI skewing (Figure 1B).
The clinical relevance of skewed XCI
Skewing of XCI can reflect or result in biological consequences for females. Because XCI is stable once established, clonal expansion of a somatic cell — as occurs in cancer — results in a cell population with extremely skewed XCI. This is often used to assess tumor clonality (7). For some X-linked diseases there is strong selection in the heterozygous carrier in favor of cells bearing the mutation on the silenced inactive X, and thus, assessment of XCI skewing in female family members can be suggestive of carrier status (8). In X-linked disorders where XCI skewing is not driven by selection, the extent of stochastic skewing can influence the extent of disease. For example, in the case of Fabry disease — an X-linked recessive inherited lysosomal storage disease — the possibility of therapeutic intervention makes the assessment of XCI skewing of increasing diagnostic value as therapies develop (9). More recently, increases in somatic XCI skewing have been associated with a variety of diagnoses, including premature ovarian failure and recurrent spontaneous abortion, several autoimmune disorders including scleroderma, and some cancers, notably breast cancer (e.g., refs. 10–12). As no causative X-linked locus has yet been identified in such associations, causes of XCI skewing in addition to selection need to be considered (e.g., refs. 10, 13).
Consideration of the sources of skewed XCI
If the assessment of XCI skewing is to be used as a clinical tool, we need to understand the biological sources of skewing. There have now been several large-scale analyses of XCI in an effort to define the basal level of skewing, the largest of which examined skewing in over 1,000 women (14). This new paper from Bolduc and colleagues (6) provides another large dataset for the literature. There is variability among studies (extreme skewing in newborns ranging from 0.5% to 2.7% of the population examined), some of which might be attributable to differences in methodology, however, the most striking changes in frequency of skewing are seen with patient age and cell type studied.
Much of the skewing seen in adult hematopoietic cells is not observed in newborns. Bolduc et al. observed a doubling of the frequency of extreme skewing in mothers relative to their newborns (6). This increase in skewing seems to become even more pronounced as women age (15). The Bolduc study also analyzed both blood and buccal (epithelial) tissue for skewing (6). They observed a substantial correlation between the two tissues, with blood showing greater skewing in both the neonates and mothers in agreement with previous studies (16, 17). This is an important finding because it suggests that tissues of different germ-lineage origin show similar inactivation patterns. As the tissue(s) affected by an X-linked disorder may be inaccessible to study, it will be important to determine the extent to which accessible tissues (such as blood or buccal samples) reflect the XCI pattern in other tissues. Age-related, acquired XCI skewing can lead to late-onset disease in individuals, as has been reported for X-linked sideroblastic anemia (18). If age-related, acquired XCI skewing occurs in tissues other than blood then late-onset symptoms might be a concern for additional X-linked disorders. Within blood, substantial differences in skewing, particularly in older adults, have been reported for different subpopulations of hematopoietic cells (19), which means that any differences in an individual’s immune health or method of sample acquisition that might bias cell representation may contribute to variability between individuals and studies.
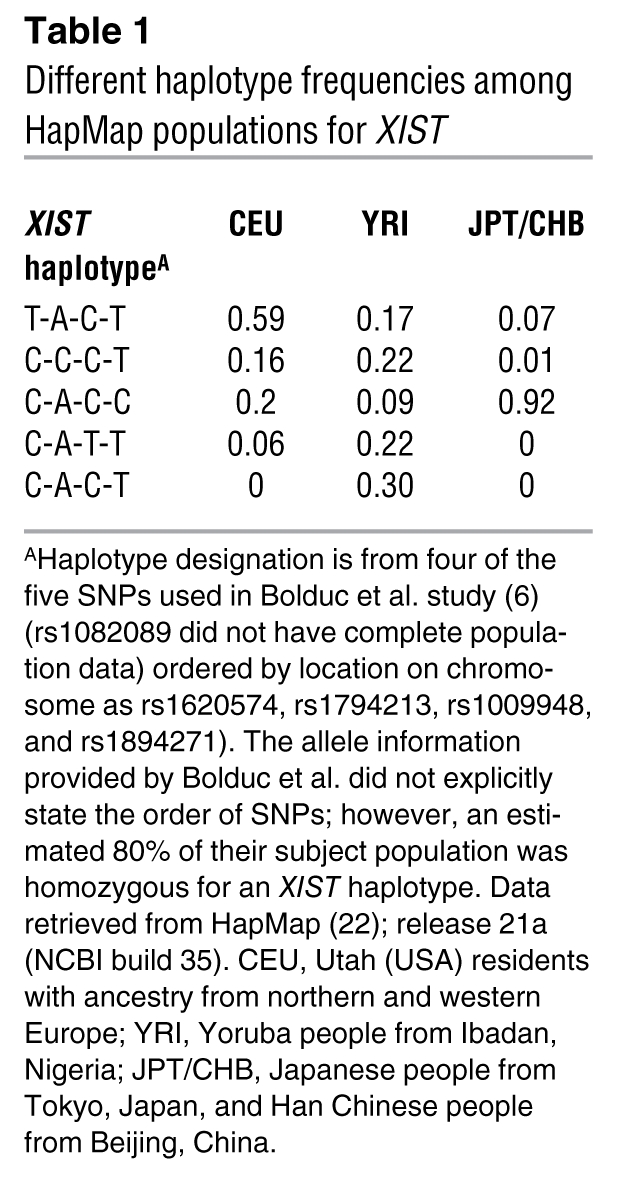
A critical question addressed by the Bolduc study was whether there is a genetic component to skewing of XCI (6). Several twin studies have shown a substantial genetic component to acquired or secondary XCI skewing (20, 21). However, there has been limited study of the intriguing question of whether there are primary influences in humans on the choice of which X chromosome to inactivate. To address this issue, Bolduc et al. examined concordance between mother-daughter pairs for XCI skewing. In both buccal and hematopoietic cells, the authors saw no evidence that neonates with skewed XCI are more likely to have had a mother with skewed XCI. There are considerable challenges to such studies as discussed by the authors. First, much of the skewing observed in the mothers could have been caused by selective pressures and acquired secondarily rather than reflecting an initial bias. Second, the low frequency of skewing in the newborn population means that sample sizes need to be very large to detect a significant heritable effect. Finally, the contribution of the paternal allele is difficult to address since males do not inactivate their only X chromosome. Therefore, drawing an analogy with the murine Xce, the authors examined polymorphisms in the XIST gene to determine whether there was a correlation between a particular allele or haplotype (combination of alleles) and the prevalence of XCI skewing. Again, no evidence for a genetic component to skewing was observed. Interestingly, however, the French-Canadian population examined in this study was relatively homogeneous for the XIST alleles studied, limiting the authors’ power to detect a significant effect. There are striking population differences between XIST haplotype frequencies (see Table 1; ref. 22), suggesting that studies in other populations (or heterogeneous populations) might reveal a different story.
Table 1 .
Different haplotype frequencies among HapMap populations for XIST
There are many case reports in the literature of families that seem to have substantial heritable skewing of XCI (for example, refs. 23, 24). Unlike in mice, in which the Xce alleles play a critical role in this skewing, such cases may reflect acquired skewing due to selection or very rare mutations related to XIST (25). While the population differences for XIST suggest we cannot yet close the book on the existence in humans of an Xce-like gene, the results from the Bolduc study in this issue of the JCI (6) support the hypothesis that the majority of XCI skewing observed in the adult population is acquired secondarily.
Acknowledgments
Work in the authors’ laboratories is supported by the Canadian Institutes of Health Research.
Footnotes
Nonstandard abbreviations used: Xce, X chromosome controlling element; XCI, X chromosome inactivation; Xist, X (inactive)–specific transcript.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 118:20–23 (2008). doi:10.1172/JCI34470.
See the related article beginning on page 333.
References
- 1.Lyon M.F. Gene action in the X-chromosome of the mouse (Mus musculus L.). . Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 2. Migeon, B.R. 2007.Females are mosaics: X inactivation and sex differences in disease. Oxford University Press. New York, New York, USA. 296 pp. [Google Scholar]
- 3.Cattanach B.M., Williams C.E. Evidence of non-random X chromosome activity in the mouse. Genet. Res. 1972;19:229–240. doi: 10.1017/s001667230001449x. [DOI] [PubMed] [Google Scholar]
- 4.Avner P., et al. Molecular correlates of the murine Xce locus. Genet. Res. . 1998;72:217–224. doi: 10.1017/s0016672398003516. [DOI] [PubMed] [Google Scholar]
- 5.Heard E., Disteche C.M. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- 6.Bolduc V., et al. No evidence that skewing of X chromosome inactivation patterns is transmitted to offspring in humans. J. Clin. Invest. 2008;118:333–341. doi: 10.1172/JCI33166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linder D., Gartler S.M. Glucose-6-phosphate dehydrogenase mosaicism: Utilization as a cell marker in the study of leiomyomas. Science. 1965;150:67–69. doi: 10.1126/science.150.3692.67. [DOI] [PubMed] [Google Scholar]
- 8.Puck J.M., Nussbaum R.L., Conley M.E. Carrier detection in X-linked severe combined immunodeficiency based on patterns of X chromosome inactivation. J. Clin. Invest. 1987;79:1395–1400. doi: 10.1172/JCI112967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobrovolny R., et al. Relationship between X-inactivation and clinical involvement in Fabry heterozygotes. Eleven novel mutations in the alpha-galactosidase A gene in the Czech and Slovak population. J. Mol. Med. 2005;83:647–654. doi: 10.1007/s00109-005-0656-2. [DOI] [PubMed] [Google Scholar]
- 10.Bretherick K., Gair J., Robinson W.P. The association of skewed X chromosome inactivation with aneuploidy in humans. Cytogenet. Genome Res. 2005;111:260–265. doi: 10.1159/000086898. [DOI] [PubMed] [Google Scholar]
- 11.Ozbalkan Z., et al. Skewed X chromosome inactivation in blood cells of women with scleroderma. . Arthritis Rheum. 2005;52:1564–1570. doi: 10.1002/art.21026. [DOI] [PubMed] [Google Scholar]
- 12.Kristiansen M., et al. High frequency of skewed X inactivation in young breast cancer patients. J. Med. Genet. 2002;39:30–33. doi: 10.1136/jmg.39.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Migeon B.R. The role of X inactivation and cellular mosaicism in women’s health and sex-specific diseases. JAMA. 2006;295:1428–1433. doi: 10.1001/jama.295.12.1428. [DOI] [PubMed] [Google Scholar]
- 14.Amos-Landgraf J.M., et al. X chromosome-inactivation patterns of 1,005 phenotypically unaffected females. Am. J. Hum. Genet. 2006;79:493–499. doi: 10.1086/507565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatakeyama C., et al. The dynamics of X-inactivation skewing as women age. Clin. Genet. 2004;66:327–332. doi: 10.1111/j.1399-0004.2004.00310.x. [DOI] [PubMed] [Google Scholar]
- 16.Sharp A., Robinson D., Jacobs P. Age- and tissue-specific variation of X chromosome inactivation ratios in normal women. Hum. Genet. 2000;107:343–349. doi: 10.1007/s004390000382. [DOI] [PubMed] [Google Scholar]
- 17.Knudsen G.P., et al. Increased skewing of X chromosome inactivation in Rett syndrome patients and their mothers. Eur. J. Hum. Genet. 2006;14:1189–1194. doi: 10.1038/sj.ejhg.5201682. [DOI] [PubMed] [Google Scholar]
- 18.Cotter P.D., et al. Late-onset X-linked sideroblastic anemia. Missense mutations in the erythroid delta-aminolevulinate synthase (ALAS2) gene in two pyridoxine-responsive patients initially diagnosed with acquired refractory anemia and ringed sideroblasts. J. Clin. Invest. 1995;96:2090–2096. doi: 10.1172/JCI118258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gale R.E., Fielding A.K., Harrison C.N., Linch D.C. Acquired skewing of X-chromosome inactivation patterns in myeloid cells of the elderly suggests stochastic clonal loss with age. . Br. J. Haematol. 1997;98:512–519. doi: 10.1046/j.1365-2141.1997.2573078.x. [DOI] [PubMed] [Google Scholar]
- 20.Kristiansen M., et al. Twin study of genetic and aging effects on X chromosome inactivation. Eur. J. Hum. Genet. 2005;13:599–606. doi: 10.1038/sj.ejhg.5201398. [DOI] [PubMed] [Google Scholar]
- 21.Vickers M.A., McLeod E., Spector T.D., Wilson I.J. Assessment of mechanism of acquired skewed X inactivation by analysis of twins. Blood. 2001;97:1274–1281. doi: 10.1182/blood.v97.5.1274. [DOI] [PubMed] [Google Scholar]
- 22.International HapMap Project. http://www.hapmap.org. [Google Scholar]
- 23.Ropers H.H., Wienker T.F., Grimm T., Schroetter K., Bender K. Evidence for preferential X-chromosome inactivation in a family with Fabry disease. Am. J. Hum. Genet. 1977;29:361–370. [PMC free article] [PubMed] [Google Scholar]
- 24.Bicocchi M.P., et al. Familial nonrandom inactivation linked to the X inactivation centre in heterozygotes manifesting haemophilia A. Eur. J. Hum. Genet. 2005;13:635–640. doi: 10.1038/sj.ejhg.5201386. [DOI] [PubMed] [Google Scholar]
- 25.Plenge R.M., et al. A promoter mutation in the XIST gene in two unrelated families with skewed X-chromosome inactivation. Nat. Genet. 1997;17:353–356. doi: 10.1038/ng1197-353. [DOI] [PubMed] [Google Scholar]