Abstract
The conversion of a normal cell to a cancer cell is a stepwise process that typically involves the activation of oncogenes and inactivation of tumour suppressor and pro-apoptotic genes1. In many instances, inactivation of genes critical for cancer development occurs by epigenetic silencing that often involves hypermethylation of CpG-rich promoter regions2,3. Whether silencing occurs by random acquisition of epigenetic marks that confer a selective growth advantage, or through a specific pathway initiated by an oncogene remains to be determined4–6. Here we perform a genome-wide RNA interference (RNAi) screen in K-ras transformed NIH 3T3 cells and identify 28 genes required for Ras-mediated epigenetic silencing of the pro-apoptotic Fas gene. At least nine of these Ras epigenetic silencing effectors (RESEs), including the DNA methyltransferase DNMT1, are directly associated with specific regions of the Fas promoter in K-ras transformed NIH 3T3 cells but not in untransformed NIH 3T3 cells. RNAi-mediated knockdown of any of the 28 RESEs results in failure to recruit DNMT1 to the Fas promoter, loss of Fas promoter hypermethylation and de-repression of Fas expression. Analysis of five other epigenetically repressed genes indicates that Ras directs silencing of multiple, unrelated genes through a largely common pathway. Finally, we show that nine RESEs are required for anchorage-independent growth and tumorigenicity of K-ras transformed NIH 3T3 cells; these nine genes have not been previously implicated in transformation by Ras. Our results demonstrate that Ras-mediated epigenetic silencing occurs through a specific, unexpectedly complex pathway involving components that are required for maintenance of a fully transformed phenotype.
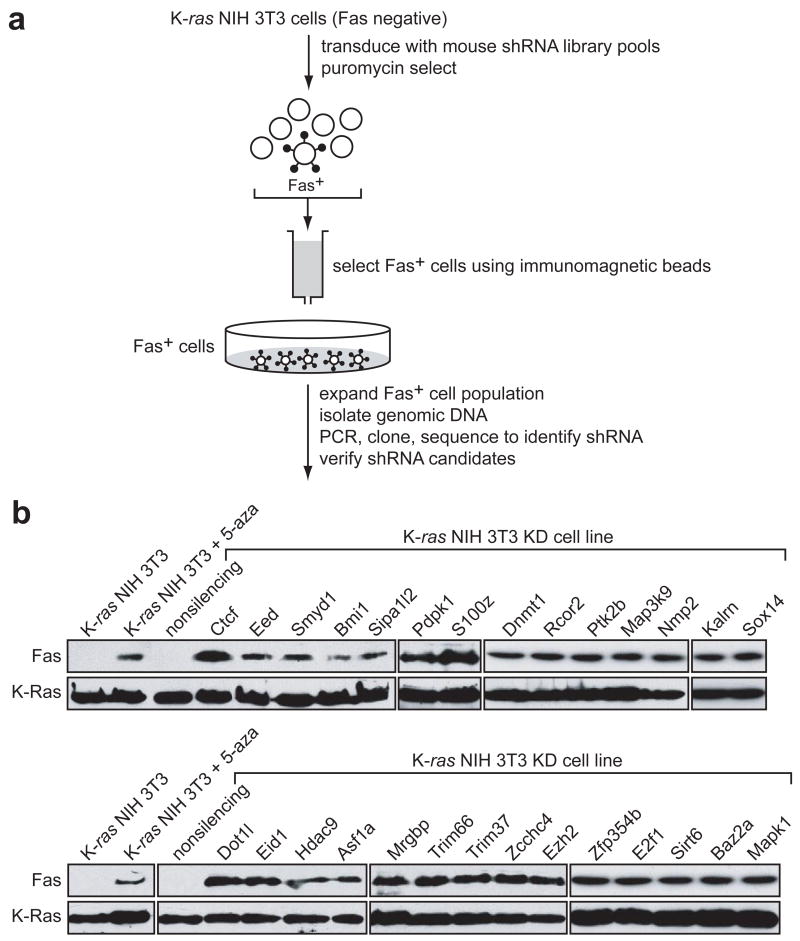
Members of the ras oncogene family transform most immortalized cell lines, and mutations of ras genes occur in ~30% of human tumours7. In addition, activation of the Ras pathway is frequent in human tumours even in the absence of ras mutations8. Previous studies have shown that in mouse NIH 3T3 cells activated Ras epigenetically silences Fas expression thereby preventing Fas-ligand induced apoptosis9,10. Activated Ras also epigenetically silences Fas expression in the human K-ras transformed cell line, HEC1A (Supplementary Fig. 1). To identify genes required for Ras-mediated silencing of Fas, we performed a genome-wide small hairpin RNA (shRNA) screen using, as a selection strategy, re-expression of Fas protein on the cell surface (Fig. 1a). A mouse shRNA library comprising ~62,400 shRNAs directed against ~28,000 genes was divided into 10 pools, which were packaged into retrovirus particles and used to stably transduce Fas-negative, K-ras NIH 3T3 cells. Fas-positive cells in each pool were selected on immunomagnetic beads using an anti-Fas antibody, the Fas-positive population was expanded, and the shRNAs were identified by sequence analysis. Positive candidates were confirmed by stably transducing K-ras NIH 3T3 cells with single shRNAs directed against the candidate genes followed by immunoblot analysis for Fas re-expression.
Figure 1. A genome-wide shRNA screen identifies factors required for Ras-mediated epigenetic silencing of Fas.
a, Schematic summary of the genome-wide shRNA screen for Ras-mediated epigenetic silencing of Fas. b, Immunoblot analysis monitoring Fas expression in the 28 K-ras NIH 3T3 knockdown (KD) cell lines. Expression of Fas in K-ras NIH 3T3 cells in the presence and absence of 5-aza-2’-deoxycytidine (5-aza) is also shown. K-Ras expression is shown as a loading control.
The screen identified 28 genes that, following shRNA-mediated knockdown, resulted in Fas re-expression. These genes are listed in Table 1 and immunoblot analysis of Fas re-expression in the 28 K-ras NIH 3T3 knockdown (K-ras NIH 3T3 KD) cell lines is shown in Fig. 1b. Consistent with previous reports10, treatment of K-ras NIH 3T3 cells with the DNA methylation inhibitor 5-aza-2’-deoxycytidine (5-aza) restored Fas expression (see also Supplementary Fig. 1). Quantitative real-time RT-PCR (qRT-PCR) confirmed in all cases that expression of the target gene was decreased in each K-ras NIH 3T3 KD cell line (Supplementary Fig. 2). For all 28 genes, a second, unrelated shRNA directed against the same target also resulted in Fas re-expression when stably expressed in K-ras NIH 3T3 cells (Supplementary Fig. 3). Knockdown of each of these 28 genes in an additional cell line, H-ras transformed murine C3H101/2 cells, also derepressed the epigenetically silenced Fas gene (Supplementary Fig. 4a).
Table 1.
Genes required for Ras-mediated epigenetic silencing of Fas
| Biological process | Accession number | Gene symbol | Name |
|---|---|---|---|
| Signal transduction | XM_993034 | Kalrn | kalirin, RhoGEF kinase |
| NM_011949 | Mapk1 | mitogen-activated protein kinase 1 | |
| NM_177395 | Map3k9 | mitogen-activated protein kinase kinase kinase 9 | |
| NM_011062 | Pdpk1 | 3-phosphoinositide dependent protein kinase 1 | |
| NM_172498 | Ptk2b | PTK2 protein tyrosine kinase 2 beta | |
| XM_193738 | S100z | S100 calcium binding protein, zeta | |
| Transcription regulation | NM_181322 | Ctcf | CCCTC-binding factor |
| NM_025613 | Eid1 | EP300 interacting inhibitor of differentiation 1 | |
| NM_007891 | E2f1 | E2F transcription factor 1 | |
| NM_054048 | Rcor2 | REST corepressor 2 | |
| XM_284529 | Sox14 | SRY-box containing gene 14 | |
| NM_181853 | Trim66 | tripartite motif-containing protein 66 | |
| NM_013744 | Zfp354b | zinc finger protein 354B | |
| Chromatin modification | NM_007552 | Bmi1 | Bmi1 polycomb ring finger oncogene |
| NM_010066 | Dnmt1 | DNA methyltransferase (cytosine-5) 1 | |
| NM_199322 | Dot1l | DOT1-like, histone H3 methyltransferase (S. cerevisiae) | |
| NM_021876 | Eed | embryonic ectoderm development | |
| NM_007971 | Ezh2 | enhancer of zeste homolog 2 (Drosophila) | |
| NM_024124 | Hdac9 | histone deacetylase 9 | |
| NM_028479 | Mrgbp | MRG binding protein | |
| NM_009762 | Smyd1 | SET and MYND domain containing 1 | |
| Chromatin Remodeling | NM_025541 | Asf1a | ASF1 anti-silencing function 1 homolog A (S. cerevisiae) |
| NM_054078 | Baz2a | bromodomain adjacent to zinc finger domain, 2A | |
| NM_181345 | Npm2 | nucleophosmin/nucleoplasmin, 2 | |
| Genome stability/Aging | NM_181586 | Sirt6 | sirtuin 6 (silent mating type information regulation 2, homolog) 6 (S. cerevisiae) |
| Unknown | XM_146572 | Sipa1l2 | signal-induced proliferation-associated gene 1 like 2 |
| NM_197987 | Trim37 | tripartite motif-containing protein 37 | |
| XM_132052 | Zcchc4 | zinc finger, CCHC domain containing 4 |
For convenience, we will refer to the protein products of the 28 genes as Ras epigenetic silencing effectors (RESEs). The RESEs include cytoplasmic cell signalling molecules and nuclear regulators of gene expression (Table 1). Among the cell signalling components, PDPK1, a serine-threonine kinase, is known to function downstream of Ras and to regulate the PI3K-AKT pathway, which is frequently activated in cancer11. Significantly, it has been previously reported that the PI3K-AKT pathway is involved in Ras-mediated silencing of Fas10. Other cell signalling proteins include MAPK1, a proximal Ras target that is frequently activated in cancer12, and PTK2B, which is recruited to cell membranes by activated Ras13.
Among the nuclear gene regulatory proteins are known transcriptional activators and repressors/corepressors (CTCF, EID1, E2F1, RCOR2, and TRIM66/TIF1D) including several predicted sequence-specific DNA binding proteins (SOX14, ZCCHC4, and ZFP345B); three histone methyltransferases (DOT1L, EZH2, and SMYD1); a histone deacetylase (HDCA9); two histone chaperones (ASF1A and NPM2); the maintenance DNA methyltransferase DNMT1; and a number of Polycomb group proteins (BMI1, EED, and EZH2). Several recent studies have linked Polycomb proteins to abnormal DNA methylation and gene silencing14–16. Surprisingly, one of the nuclear RESEs is BAZ2A/TIP5, previously known only to be involved in repression of RNA polymerase I-directed ribosomal gene transcription17. A number of RESEs were substantially upregulated at the transcriptional (Supplementary Fig. 5) or post-transcriptional (Supplementary Fig. 6) level in K-ras NIH 3T3 cells compared to NIH 3T3 cells, explaining, at least in part, how K-ras activates this silencing pathway.
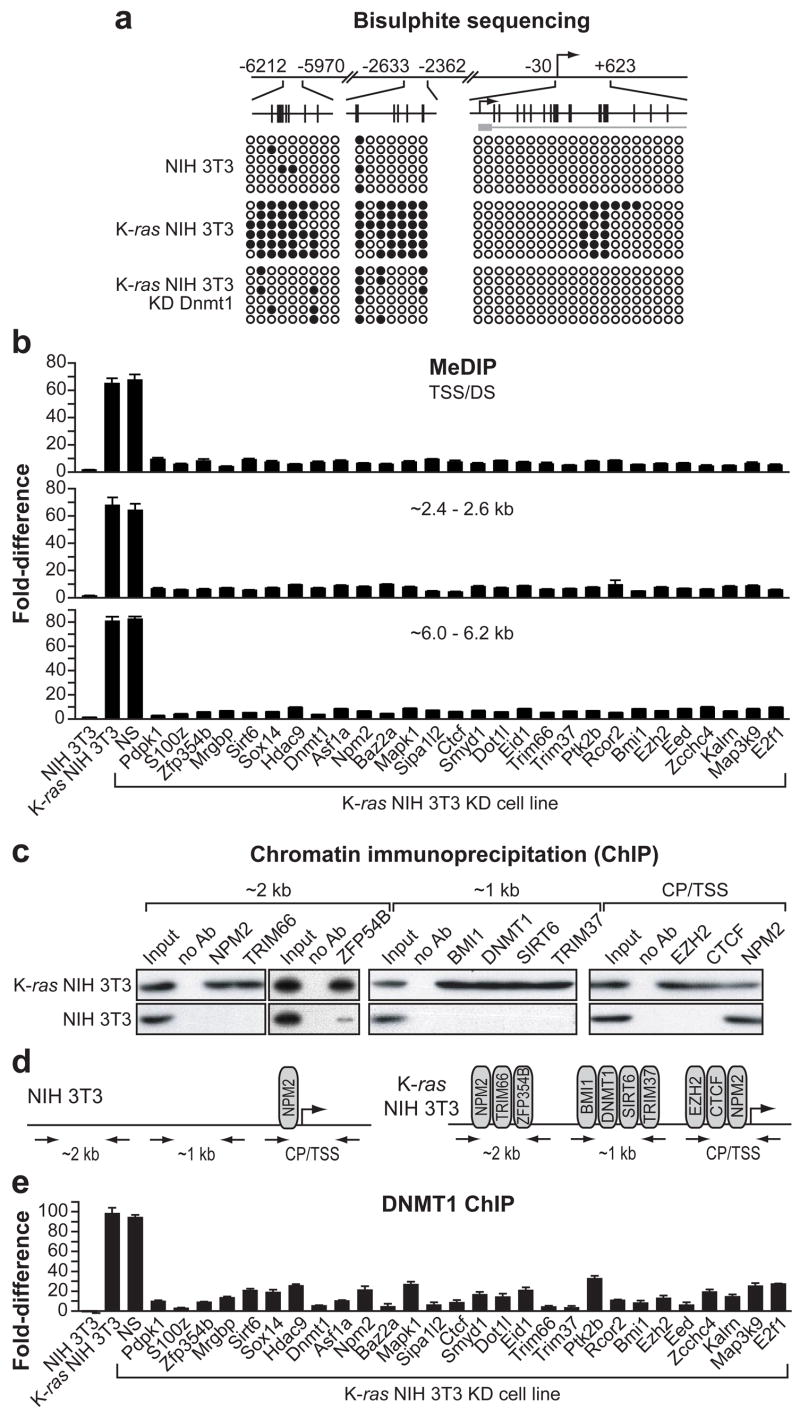
As mentioned above, treatment of K-ras NIH 3T3 cells with 5-aza results in Fas re-expression, suggesting that repression is due, at least in part, to promoter hypermethylation. We therefore sought to determine the relationship between Fas promoter hypermethylation and Fas re-expression following knockdown of each of the 28 RESEs. We first confirmed that the repressed Fas promoter was hypermethylated and mapped the hypermethylated region(s) by bisulphite sequence analysis. These results, summarized in Fig. 2a, reveal three regions located upstream and downstream from the transcription start-site (TSS) that are hypermethylated in K-ras NIH 3T3 cells but not in NIH 3T3 cells or in K-ras NIH 3T3 cells following knockdown of DNMT1. Significantly, these same three Fas promoter regions are also hypermethylated in H-ras transformed C3H10T1/2 cells but not in C3H10T1/2 cells or in H-ras transformed C3H10T1/2 cells following knockdown of DNMT1 (Supplementary Fig. 4b).
Figure 2. ChIP analysis and methylation status of the Fas promoter.
a, Summary of bisulphite sequencing. Open circle, unmethylated CpG; closed circle, methylated CpG. Each row represents a single clone. CpG dinucleotide positions are shown by vertical lines. b, MeDIP analysis following knockdown of each of the 28 RESEs. NS, nonsilencing shRNA. Values are expressed as the fold-difference relative to input, and have been corrected for background. Error bars indicate standard error (n=3). c, ChIP assay monitoring occupancy of selected RESEs at the core promoter/TSS (CP/TSS) or ~1 kb or ~2 kb upstream of the TSS. d, Summary of the ChIP results. e, ChIP analysis monitoring occupancy of DNMT1.
To facilitate analysis of the methylation status of these three regions in the 28 K-ras NIH 3T3 KD cell lines we established and validated a rapid methylated DNA immunoprecipitation (MeDIP) assay, in which the antibody is directed against 5-methyl-cytosine18. The MeDIP results of Supplementary Fig. 7 show that in K-ras NIH 3T3 cells the Fas promoter was hypermethylated within the TSS/downstream (DS) region, consistent with the bisulphite sequencing results. Moreover, the MeDIP results show, as expected, that the TSS/DS region was not hypermethylated in NIH 3T3 cells or in K-ras NIH 3T3 cells following 5-aza treatment. We then assessed the three hypermethylated Fas promoter regions in each of the 28 K-ras NIH 3T3 KD cell lines. The results of Fig. 2b show that in all 28 K-ras NIH 3T3 KD cell lines the three Fas promoter regions were not hypermethylated, consistent with the expression data.
To further understand the basis of Fas silencing, we asked whether nuclear RESEs functioned by direct association with the Fas promoter. We performed a series of chromatin immunoprecipitation (ChIP) assays, based upon antibody availability, using three sets of promoter-specific primer-pairs located ~2 kb upstream of the TSS, ~1 kb upstream of the TSS or encompassing the core promoter/TSS. The three primer-pairs cover the entire Fas promoter region. Figure 2c shows that in K-ras NIH 3T3 cells, nine of the RESEs were bound to specific Fas promoter regions: NPM2, TRIM66 and ZFP354B were present ~2 kb upstream of the TSS; BMI1, DNMT1, SIRT6 and TRIM37 were present ~1 kb upstream of the TSS; and EZH2, CTCF and NPM2 were present at the core promoter/TSS. Significantly, in NIH 3T3 cells only NPM2 was detectably associated with the Fas promoter at the core promoter/TSS. Thus, as summarized in Fig. 2d, at least nine RESEs are recruited to specific regions of the Fas promoter in response to expression of activated Ras.
The ChIP results of Fig. 2c show that DNMT1 is associated with the Fas promoter in K-ras NIH 3T3 cells but not in untransformed NIH 3T3 cells. The two other DNA methyltransferases, DNMT3A and DNMT3B, were not identified in the original shRNA screen and are not detectably associated with the Fas promoter by ChIP analysis (Supplementary Fig. 8). These results strongly suggest that DNMT1 is required to sustain hypermethylation of the Fas promoter in K-ras NIH 3T3 cells. To confirm this possibility, we analyzed association of DNMT1 with the Fas promoter in the 28 K-ras NIH 3T3 KD cell lines. The ChIP results of Fig. 2e show that in all 28 K-ras NIH 3T3 KD cell lines association of DNMT1 with the Fas promoter was markedly reduced. Moreover, bisulphite sequence analysis showed that following knockdown of DNMT1 the TSS/DS region of the Fas promoter was no longer hypermethylated (see Fig. 2a).
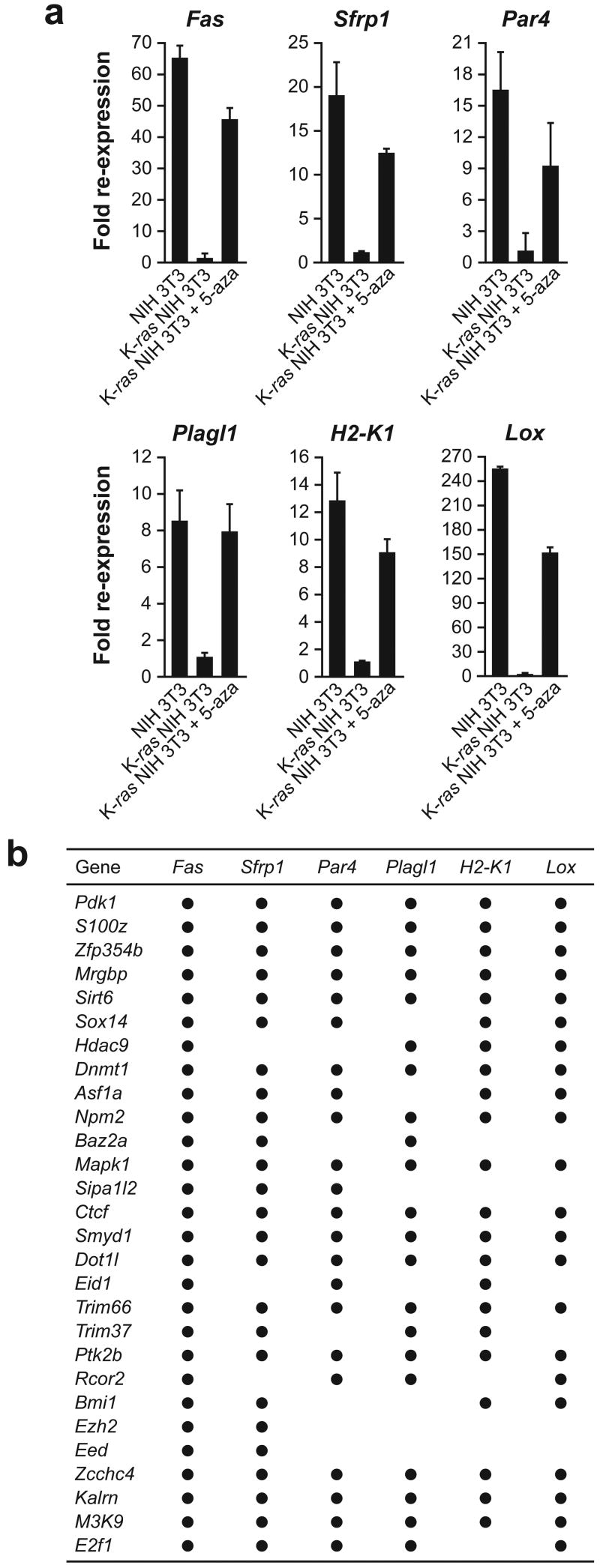
A number of genes in addition to Fas are known to be epigenetically silenced in ras transformed cells. To gain insight into whether Ras mediates epigenetic silencing of different genes through common or diverse pathways, we analyzed five other well-studied, epigenetically silenced genes: Sfrp1, Par4/Pawr, Plagl1, H2-K1 and Lox. A variety of evidence supports the relevance of these genes to cellular transformation and cancer (reviewed in refs. 19–23). The results of Fig. 3a show that, like Fas, all five genes were expressed in NIH 3T3 cells but not in K-ras NIH 3T3 cells, and were re-expressed in K-ras NIH 3T3 cells following treatment with 5-aza. Bisulphite sequence analysis confirmed that all five genes contained regions that are hypermethylated in K-ras NIH 3T3 cells but not in NIH 3T3 cells or in K-ras NIH 3T3 cells following knockdown of DNMT1 (Supplementary Fig. 9). For four of these genes (Sfrp1, Par4, Plagl1 and H2-K1), the TSS was encompassed by dense hypermethylation in K-ras NIH 3T3 cells.
Figure 3. Ras directs epigenetic silencing of multiple, unrelated genes through a largely common pathway.
a, qRT-PCR monitoring expression of Fas, Sfrp1, Par4, Plagl1, H2-K1 and Lox in NIH 3T3 cells, and in K-ras NIH 3T3 cells in the presence and absence of 5-aza. Values are expressed as fold re-expression relative to expression of the gene in K-ras NIH 3T3 cells, which is arbitrarily set to 1. Error bars indicate standard error (n=3). b, Summary of qRT-PCR analysis monitoring re-expression of Fas, Sfrp1, Par4, Plagl1, H2-K1 and Lox following knockdown of each of the 28 RESEs.
We next analyzed expression of Sfrp1, Par4, Plagl1, H2-K1 and Lox in the 28 K-ras NIH 3T3 KD cell lines. The qRT-PCR results of Supplementary Fig. 10 are summarized in Fig. 3b and reveal substantial overlap in the requirements of RESEs for epigenetic silencing of Fas, Sfrp1, Par4, Plagl1, H2-K1 and Lox: of the 28 RESEs required for silencing of Fas, at least 21 were also required for silencing of each of the five other genes analyzed. MeDIP analysis for all five genes revealed a perfect correspondence between the RESEs required for silencing and for promoter hypermethylation (Supplementary Fig. 11). These results indicate that Ras directs the epigenetic silencing of multiple, unrelated genes through a largely common pathway.
Proteins that function downstream of Ras could be essential for a fully transformed phenotype. To determine whether any of the 28 RESEs were also required for Ras-mediated transformation, we first tested the ability of the K-ras NIH 3T3 KD cell lines to grow in soft agar. Supplementary Fig. 12a shows that knockdown of any of nine RESEs (S100Z, MRGBP, BAZ2A, SMYD1, EID1, TRIM66, TRIM37, ZCCHC4, and KALRN) markedly inhibited anchorage-independent growth.
To further characterize the role of these nine RESEs in Ras-mediated transformation, we tested the ability of the corresponding nine K-ras NIH 3T3 KD cell lines to form tumours following subcutaneous injection in the flanks of nude mice. Supplementary Fig. 12b show that knockdown of SMYD1 or BAZ2A moderately inhibited tumour growth, whereas knockdown of S100Z, TRIM37, TRIM66, EID1, ZCCHC4, MRGBP, or KALRN markedly inhibited tumour growth.
It is well established that in many cancers specific genes affecting cellular growth control are hypermethylated and epigenetically silenced2,3. However, the mechanistic basis of epigenetic silencing is not understood. The results presented here demonstrate that oncogenic Ras directs epigenetic silencing through a specific, unexpectedly complex pathway involving at least 28 components (RESEs). Knockdown of any of the 28 RESEs resulted in a failure to recruit DNMT1 to the Fas promoter, loss of Fas promoter hypermethylation and de-repression of Fas expression. The vast majority of RESEs have not been previously connected to the Ras pathway, and thus our results have identified a number of new factors that act downstream of Ras. Nine RESEs are required for anchorage-independent growth and tumorigenicity. The identification of new components that act downstream of Ras, and are required for epigenetic silencing and complete transformation, provides potential new anti-cancer targets.
METHODS SUMMARY
shRNA screen
The mouse shRNAmir library (release 2.16; Open Biosystems) was used to generate ten retroviral pools, each comprising ~6000 shRNA clones. K-ras NIH 3T3 cells were transduced with the retroviral pools, and 2 days later selected for resistance to puromycin for 7 days. Fas-positive cells were selected, expanded and genomic DNA isolated. To identify the candidate shRNAs, the shRNA region of the transduced virus was PCR amplified, cloned and sequenced.. Individual knockdown cell lines were generated by retroviral transduction of K-ras NIH 3T3 cells with the respective shRNA. Individual shRNAs were either obtained from the Open Biosystems library or synthesized (see Supplementary Table 1).
Bisulphite sequencing
Bisulphite modification was carried out essentially as described24 except that hydroquinone was used at a concentration of 125 mM during bisulphite treatment carried out in the dark and DNA was desalted on Qiaquick columns (Qiagen) after the bisuphite reaction.
Chromatin immunoprecipitation (ChIP) and methylated DNA immunoprecipitation (MeDIP)
ChIP assays were performed using extracts prepared 7 days following retroviral transduction and puromycin selection. The sequences of the primers used for amplifying the MeDIP and ChIP products are provided in Supplementary Table 2.
Quantitative real time RT-PCR
Total RNA was isolated and reverse transcription performed, followed by quantitative real-time PCR. Primer sequences are provided in Supplementary Table 2.
Soft agar assays
Soft agar assays were performed using the CytoSelect 96-well Cell Transformation Assay (Cell Biolabs) as per the manufacturer’s instructions.
Tumour formation assays
5×106 NIH 3T3, K-ras NIH 3T3, or K-ras NIH 3T3 knockdown Cells were injected subcutaneously into the right flank of athymic Balb/c (nu/nu) mice (Taconic). Tumour dimensions were measured every 3 days from the time of appearance of the tumours. Animal experiments were performed in accordance with the Institutional Animal Care and Use Committee (IACUC) guidelines.
METHODS
Cell culture
NIH 3T3 (ATCC# CRL-1658) and K:Molv NIH 3T3 (ATCC# CRL-6361; referred to here as K-ras NIH 3T3) cells were maintained in DMEM supplemented with 10% FCS at 37°C and 5% CO2. For 5-aza-2’-deoxycytidine (5-aza) treatment, K-ras NIH 3T3 cells were treated with 10 μM 5-aza for 72 h.
shRNA screen
The mouse shRNAmir library (release 2.16; Open Biosystems) was obtained through the University of Massachusetts Medical School shRNA library core facility. Ten retroviral pools, each comprising ~6000 shRNA clones, were generated with titers of ~2.6×105 pfu ml−1. These retroviral stocks were produced following co-transfection into the PhoenixGP packaging cell line (a gift from G. Nolan, Stanford University, USA). K-ras NIH 3T3 cells (1.2×106) were transduced at an MOI of 0.2 with the retroviral stocks in 100 mm plates, and 2 days later selected for resistance to puromycin (1.5 μg ml−1) for 7 days. To isolate Fas-positive cells, 5×106 cells from each pool were incubated with an anti-Fas antibody (15A7; eBiosciences) followed by incubation with IgG-conjugated magnetic beads (Miltenyi Biotec), and Fas-positive cells were selected using the Mini MACS magnetic separation system (Miltenyi Biotec) according to the manufacturer’s instructions. The selected Fas-positive cells were expanded and genomic DNA isolated. To identify the candidate shRNAs, the shRNA region of the transduced virus was PCR amplified (using primers PSM2-forward, 5’-GCTCGCTTCGGCAGCACATATAC-3’ and PSM2-reverse, 5’-GAGACGTGCTACTTCCATTTGTC-3’) and cloned into pGEM-T Easy (Promega). An average of 30 clones were sequenced per pool (using primer PSM2-seq, 5’-GAGGGCCTATTTCCCATGAT-3’). Individual knockdown cell lines were generated by retroviral transduction of 0.6×105 K-ras NIH 3T3 cells with the respective shRNA. Individual shRNAs were either obtained from the Open Biosystems library or synthesized (see Supplementary Table 1).
Immunoblot analysis
To prepare cell extracts, K-ras NIH 3T3 knockdown cell lines were harvested 7 days following retroviral transduction and puromycin selection (1.5 μg ml−1) and lysed by boiling in 1X SDS sample buffer (Laemmli buffer) for 5 min. Proteins were resolved by 12% SDS-PAGE. Immunoblot analysis was performed using an anti-Fas (sc-716; Santa Cruz) or anti-p21 Ras (ab16795; Abcam) antibody to monitor expression of K-Ras (as a loading control), and an appropriate HRP-conjugated secondary antibody. Proteins were visualized using SuperSignal West Pico Luminol/Enhancer Solution (Pierce).
Bisulphite sequencing
For each cell line six clones were sequenced. The regions analyzed were amplified by nested PCR. The first round comprised 24 cycles at 94°C for 1 min, 48°C for 1 min 30 s, and 72°C for 1 min. One-tenth of the product was used as substrate for the second round of PCR comprising 28 cycles at 94°C for 1 min, 48°C for 1 min 30 s, 72°C for 1 min. Primer sequences are provided in Supplementary Table 2.
Chromatin immunoprecipitation (ChIP) and methylated DNA immunoprecipitation (MeDIP)
The following antibodies were used: anti-5-methyl cytosine (ab1884; Abcam), anti-EZH2 (4905; Cell Signaling Technology), anti-CTCF (07-729; Upstate), anti-BMI1 (ab14389; Abcam), anti-DNMT1 (IMG-261A; Imgenex), anti-SIRT6 (ASB-ARP32409; Aviva Systems Biology), anti-TRIM37 (a gift from A.E. Lehesjoki, Folkhalsan Institute of Genetics, Finland), anti-TRIM66 (a gift from R. Losson, IGBMC, France), and anti-NPM2 (a gift from M.M. Matzuk, Baylor College of Medicine, USA). The anti-ZFP354B antibody was raised against a synthetic peptide corresponding to amino acids 126-143 of the murine protein, and affinity purified on a peptide coupled to agarose. For ChIP analysis of the Fas promoter, primer-pairs located at the core promoter/TSS, ~1 kb upstream of the TSS or ~2 kb upstream of the TSS were used for PCR analysis of the input and immunoprecipitated DNA samples. MeDIP and ChIP products were visualized by autoradiography, or analyzed by quantitative real-time PCR using Platinum SYBR Green qPCR SuperMix-UDG with Rox (Invitrogen). Calculation of fold-differences was done as previously described25.
Quantitative real time RT-PCR
Total RNA was isolated using TRIZOL (Invitrogen) 7 days following retroviral transduction and puromycin selection. Reverse transcription was performed using SuperScript II Reverse Transcriptase (Invitrogen) as per the manufacturer’s instructions, followed by quantitative real-time PCR as described above. Primer sequences are provided in Supplementary Table 2.
Tumour formation assays
5×106 NIH 3T3, K-ras NIH 3T3, or K-ras NIH 3T3 knockdown cell lines were suspended in 100 μl of serum-free DMEM and injected subcutaneously into the right flank of athymic Balb/c (nu/nu) mice (Taconic). Tumour dimensions were measured every 3 days from the time of appearance of the tumours, and tumour volume was calculated using the formula π/6 × (length) × (width)2. Animal experiments were performed in accordance with the Institutional Animal Care and Use Committee (IACUC) guidelines.
Supplementary Material
Supplementary Information is linked to the online version of the paper on www.nature.com/nature.
Acknowledgments
We thank M. Koken, A. E. Lehesjoki, R. Losson, M. M. Matzuk, G. Nolan, E. J. Taparowsky and T. A. Waldman for reagents, and S. Evans for editorial assistance. C.G. is on leave from the CNRS, Paris, France. This work was funded in part by a grant from the NIH to M.R.G. M.R.G. is an investigator of the Howard Hughes Medical Institute.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2(Suppl 1):S4–11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 3.Esteller M. Epigenetics provides a new generation of oncogenes and tumour-suppressor genes. Br J Cancer. 2006;94:179–183. doi: 10.1038/sj.bjc.6602918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones PA. DNA methylation errors and cancer. Cancer Res. 1996;56:2463–2467. [PubMed] [Google Scholar]
- 5.Baylin S, Bestor TH. Altered methylation patterns in cancer cell genomes: cause or consequence? Cancer Cell. 2002;1:299–305. doi: 10.1016/s1535-6108(02)00061-2. [DOI] [PubMed] [Google Scholar]
- 6.Keshet I, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 7.Giehl K. Oncogenic Ras in tumour progression and metastasis. Biol Chem. 2005;386:193–205. doi: 10.1515/BC.2005.025. [DOI] [PubMed] [Google Scholar]
- 8.Ehmann F, et al. Detection of N-RAS and K-RAS in their active GTP-bound form in acute myeloid leukemia without activating RAS mutations. Leuk Lymphoma. 2006;47:1387–1391. doi: 10.1080/10428190600565925. [DOI] [PubMed] [Google Scholar]
- 9.Fenton RG, Hixon JA, Wright PW, Brooks AD, Sayers TJ. Inhibition of Fas (CD95) expression and Fas-mediated apoptosis by oncogenic Ras. Cancer Res. 1998;58:3391–3400. [PubMed] [Google Scholar]
- 10.Peli J, et al. Oncogenic Ras inhibits Fas ligand-mediated apoptosis by downregulating the expression of Fas. EMBO J. 1999;18:1824–1831. doi: 10.1093/emboj/18.7.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osaki M, Oshimura M, Ito H. PI3K-Akt pathway: its functions and alterations in human cancer. Apoptosis. 2004;9:667–676. doi: 10.1023/B:APPT.0000045801.15585.dd. [DOI] [PubMed] [Google Scholar]
- 12.de Vries-Smits AM, Burgering BM, Leevers SJ, Marshall CJ, Bos JL. Involvement of p21ras in activation of extracellular signal-regulated kinase 2. Nature. 1992;357:602–604. doi: 10.1038/357602a0. [DOI] [PubMed] [Google Scholar]
- 13.Alfonso P, et al. Proteomic analysis of p38alpha mitogen-activated protein kinase-regulated changes in membrane fractions of RAS-transformed fibroblasts. Proteomics. 2006;6(Suppl 1):S262–271. doi: 10.1002/pmic.200500350. [DOI] [PubMed] [Google Scholar]
- 14.Ohm JE, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlesinger Y, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 16.Widschwendter M, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Y, Santoro R, Grummt I. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 2002;21:4632–4640. doi: 10.1093/emboj/cdf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 19.Rubin JS, Barshishat-Kupper M, Feroze-Merzoug F, Xi ZF. Secreted WNT antagonists as tumor suppressors: pro and con. Front Biosci. 2006;11:2093–2105. doi: 10.2741/1952. [DOI] [PubMed] [Google Scholar]
- 20.Ranganathan P, Rangnekar VM. Regulation of cancer cell survival by Par-4. Ann N Y Acad Sci. 2005;1059:76–85. doi: 10.1196/annals.1339.046. [DOI] [PubMed] [Google Scholar]
- 21.Abdollahi A. LOT1 (ZAC1/PLAGL1) and its family members: mechanisms and functions. J Cell Physiol. 2007;210:16–25. doi: 10.1002/jcp.20835. [DOI] [PubMed] [Google Scholar]
- 22.Nie Y, et al. DNA hypermethylation is a mechanism for loss of expression of the HLA class I genes in human esophageal squamous cell carcinomas. Carcinogenesis. 2001;22:1615–1623. doi: 10.1093/carcin/22.10.1615. [DOI] [PubMed] [Google Scholar]
- 23.Kenyon K, et al. Lysyl oxidase and rrg messenger RNA. Science. 1991;253:802. doi: 10.1126/science.1678898. [DOI] [PubMed] [Google Scholar]
- 24.Frommer M, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper on www.nature.com/nature.