Abstract
Adeno-associated virus (AAV)-based vectors have been shown to be effective in transferring the cystic fibrosis gene (CFTR) into airway epithelial cells in animal models and in patients. However, the level of CFTR gene expression has been low because the vector cannot accommodate the CFTR gene together with a promoter. In this study, we described a strategy to reduce the size of the CFTR cDNA to allow the incorporation of an effective promoter with the CFTR gene into AAV vectors. We engineered and tested 20 CFTR mini-genes containing deletions that were targeted to regions that may contain nonessential sequences. Functional analyses showed that four of the shortened CFTRs (one with combined deletions) retained the function and the characteristics of a wild-type CFTR, as measured by open probability, time voltage dependence, and regulation by cAMP. By using an AAV vector with a P5 promoter, we transduced these short forms of CFTR genes into target cells and demonstrated high levels of CFTR expression. We also demonstrated that smaller AAV/CFTR vectors with a P5 promoter expressed the CFTR gene more efficiently than larger vectors or a vector in which CFTR gene was expressed from the AAV inverted terminal repeat sequence. The CFTR mini-gene with combined deletions was packaged into AAV virions more efficiently, generated higher titers of transducing virions, and more effectively transferred CFTR function into target cells. These new vectors should circumvent the limitations of AAV vector for CFTR expression. Our strategy also may be applicable to other genes, the sizes of which exceed the packaging limit of an AAV vector.
Keywords: cystic fibrosis/gene therapy/deletions/patch clamp/chloride channel
Cystic fibrosis (CF) is a common autosomal recessive disease caused by mutations in the gene encoding the cystic fibrosis transmembrane conductance regulator (CFTR) (1, 2). The CFTR gene encodes a cAMP-regulated Cl− channel that functions on the apical surface of epithelial cells (3, 4). Although CF affects multiple systems, lung disease is the major cause of morbidity. Studies of the epithelial cells from patients with CF have shown that mutations of CFTR gene cause defective regulation of Cl− permeability across apical membranes (5–8). This defect is believed to be responsible for the disease in the lung, where the deficiency of Cl− reabsorption in airway epithelia is associated with hypertonic secretion that causes the inactivation of defensins, a family of salt-sensitive proteins that inhibits microbes in the lung airways (9). As a consequence, bacteria, mainly pseudomonas, colonize the airways and induce inflammatory response, resulting in thick purulent mucus and progressive loss of airway function (10, 11).
Like most of the human genetic diseases, CF requires life-long therapeutic interventions, and the median survival of the patients is ≈26 years. Gene therapy is a promising alternative for treating CF because it has the potential to provide long term relief by expressing the normal CFTR gene in the diseased tissues. The feasibility of gene transfer to lung cells initially was demonstrated by the findings that expression of the wild-type CFTR cDNA corrected the chloride channel defect in vitro using retrovirus or adenovirus vectors (12, 13). However, gene therapy of CF in patients has not been successful because of the limitations of the current vector systems when used in vivo.
The ideal gene therapy regimen is one that allows stable maintenance of the wild-type gene inside the defective cells and provides long term therapeutic effects within the diseased tissue without serious side effects. Adenoviral vectors can infect airway epithelia efficiently (14–18). However, these vectors are not maintained stably within the cells and have been shown to cause immune and inflammatory responses in animal models and in patients (19–22) due to the expression of the viral genes carried by the vectors (23).
Recombinant adeno-associated virus (rAAV) has attracted increasing attention as a vector for gene therapy of CF because it naturally infects the airway epithelial cells and has not been associated with any disease. Vectors derived from AAV can infect nondividing cells and provide prolonged gene expression. In addition, AAV vectors deliver therapeutic genes without cotransfer of any viral genes and therefore are not likely to induce inflammatory responses. Indeed, experiments with rAAV-mediated gene transfer into skeletal muscles have shown transgene expression for 1 year or more with no inflammatory responses in animal models (24–26).
For the study of gene therapy of CF, rAAV has been shown to infect airway cells efficiently, and the vector DNA persisted for weeks in animal models (27, 28). Recent phase I clinical trials also have shown efficient delivery of the CFTR gene into epithelial cells and significant improvements in isoproterenol-induced chloride currents judged by potential difference recordings in maxillary sinus of patients with CF (29, 30). The delivered CFTR cDNA persisted over 70 days based on PCR detection of vector DNA. The ongoing trial at Johns Hopkins University (29) also has shown AAV vector administration into lungs to be safe and well tolerated in patients.
Despite these advantages and promising results, the levels of CFTR expression from AAV vector may be suboptimal because the size of the coding sequence of the CFTR cDNA is larger than the optimal size that can be packaged into an AAV vector. Addition of an effective promoter will further compromise the packaging efficiency. Thus, in the current AAV/CFTR vector under clinical trials, the CFTR gene is driven by an AAV inverted terminal repeat (ITR) sequence, where a putative promoter like-activity was identified (28, 31). However, this ITR promoter has a very low transcriptional activity compared with conventional promoters usually used to drive gene expression.
The optimal size range for AAV packaging is between 4.0 and 4.8 kb (32). Although AAV can package a vector up to 5.0 kb, the packaging efficiency decreases sharply with increasing vector size. The essential sequences for an AAV vector are the two ITRs, each 145 bp in size. Therefore, AAV vector can accommodate a transcriptional cassette, including promoter and polyadenylation signal, up to 4.7 kb in size. Because the CFTR cDNA to the stop codon gene is 4.6 kb and a poly(A) signal is also essential, it has not been possible to incorporate an additional promoter with the CFTR cDNA into the AAV vector.
In this report, we describe a strategy for reducing the size of CFTR cDNA by generating deletions in the coding region that do not compromise its function. When delivered with an AAV vector, these shortened genes impart high levels of Cl− channel activity. This strategy provides a solution to the limitations of AAV as a vector for gene therapy of CF.
MATERIALS AND METHODS
Cells and Vectors.
HeLa and 293 cells were obtained from the American Type Culture Collection. The cells were maintained in DMEM (GIBCO) supplemented with 10% fetal bovine serum (GIBCO).
AAV vectors containing the shortened CFTR genes were constructed by first inserting the CFTR cDNA into pP5TK65, which contains the P5 promoter followed by a 65-bp, truncated polyadenylation [poly(A)] signal from the TK gene of herpes simplex virus. The transcriptional cassette containing the CFTR gene then was inserted into an AAV vector, pAV53 (unpublished work). The two ITR sequences of pAV53 and part of the linker sequences were 302 bp in total length. The P5 promoter we used was 150 bp in length. For comparing the levels of expression, a minimal CFTR cDNA (S2) containing the entire CFTR coding sequence was cloned into pAV53 with only the TK poly(A) signal. In this construct, the CFTR cDNA is expressed from the putative promoter in the ITR sequence. Transducing AAV virions were prepared as described (33).
Reduction in the Coding Region of CFTR cDNA.
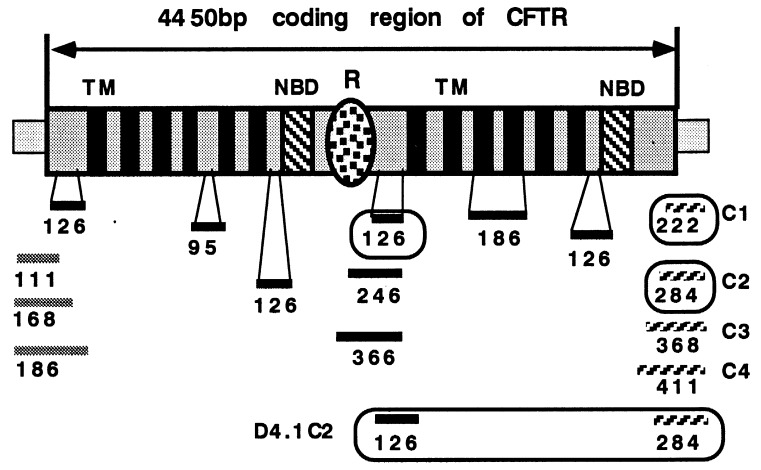
The CFTR cDNA is >7.2 kb, of which the sequence encoding for the wild-type CFTR protein is 4.45 kb. However, to express efficiently the mini-gene, it requires some untranslated regions. Hence, in our construct, 30-bp containing the Kozak sequences were included upstream of the AUG codon, and 65 bp containing the poly(A) signal were included downstream of the stop codon of the CFTR cDNA. To reduce further the size of the coding region, we targeted deletions into regions that have probability of containing nonessential sequences. Deletions were introduced into the CFTR cDNA by using a two-step PCR amplification. The first 5′-primer contained sequences flanking the regions to be deleted. The 3′-primer spanned or passed a unique restriction site that was to be used for cloning the amplified fragment. After the first amplification, a second 5′-primer that spanned a second restriction at the 5′-end of the fragment was used for the second amplification. The final amplified fragments were sequenced and cloned into the corresponding region of the CFTR in a plasmid, pBQ4.5, which contained the entire coding region of CFTR and the minimal untranslated regions. We made 20 deletions ranging from 51 to 360 bp at the N-terminal, the cytoplasmic tail region, the transmembrane domains, the loop regions between transmembrane domains, and the R domain. The regions into which we have introduced deletions are listed in Table 1.
Table 1.
Deletions in the CFTR mini gene
| Constructs* | Regions deleted† | Nucleotides deleted‡ | aa removed, n |
|---|---|---|---|
| ΔC1 | C | 4,503–4,725 | 23 |
| ΔC2 | C | 4,441–4,725 | 44 |
| ΔC3 | C | 4,357–4,725 | 72 |
| ΔC4 | C | 4,314–4,725 | 86 |
| ΔN1 | N | 73–184 | 17 |
| ΔN2 | N | 73–241 | 36 |
| ΔN3 | N | 73–295 | 54 |
| D1 | N | 247–372 | 42 |
| ΔT2 | N | 256–646 | 130 |
| ΔT4 | N | 256–922 | 222 |
| ΔT6 | N | 298–1,342 | 348 |
| D2 | L | 886–981 | 32 |
| D3 | L | 1,225–1,351 | 42 |
| D4.1 | R | 2,521–2,647 | 42 |
| D4.2 | R | 2,401–2,647 | 82 |
| D4.3 | R | 2,281–2,647 | 122 |
| D5 | T | 3,058–3,244 | 62 |
| D6 | L | 3,601–3,727 | 42 |
| D4.1C1 | LC | 2,521–2,647/ | 65 |
| 4,503–4,725 | |||
| D4.1C2 | LC | 2,521–2,647/ | 86 |
| 4,441–4,725 |
ΔC1, ΔC2, ΔC3, and ΔC4 contain stop codons from the primers that are used to make the deletions. ΔN1, ΔN2, and ΔN3 have the Kozak sequences from the primers used.
C, N, L, or R indicates C-terminal, N-terminal, loop region, and regulation domain, respectively.
The numbers of nucleotides deleted in C1-4 and N1-3 deletions include nontranslated regions. In the wild-type cftr cDNA, the start codon and the stop codon are located at nt 133 and nt 4,573, respectively (1).
Assay for Promoter Strength.
Promoter strengths were compared by using a chloramphenicol acetyltransferase (CAT) gene assay as described (32). In brief, CMVie, SV40e, or AAV P5 promoter was inserted with a CAT expression cassette in an AAV vector. These vectors were packaged into virions under identical conditions. The promoter strengths were determined by the amounts of CAT activity in HeLa cells that had been infected with the same volumes of each viral lysate.
Functional Assays of the Shortened CFTR.
Initial screening for functional shortened CFTR was carried out by using an established 125I-efflux assay (34). In brief, 293 cells were transfected stably with a plasmid containing each of the shortened CFTR mini-cDNA. The cells grown on 18-mm cover glass (Fisher) in 24-well plates (Nunc) were incubated in culture medium containing Na125I (20 μCi/ml) for 1 h. Incubation medium was removed, and the cells were washed twice briefly (10 s) with Kreb’s buffer (34) to remove extracellular Na125I. Medium (1 ml) then was added to the cells, removed, and replaced with fresh medium at 1-min intervals. Cells then were stimulated with 10 mM forskolin, 1 mM isobutylmethylxanthine, and 200 mM 8-(4-chlorophenylthio)–CAMP with additional medium changes and incubations. Cells were lysed in 0.1 N NaOH (1 ml), and radioactivity in cell and efflux samples was measured with a γ counter. The effluxes were expressed as fractional rate of loss = [(A − B)/A] × 100, where A is the number of counts in the cells at the start of a 1-min efflux period and B is the counts at the end.
Whole-cell patch–clamp was used to analyze the levels of CFTR expression from the AAV vectors in transduced HeLa cells. The characteristics of the shortened CFTR genes also were analyzed with single-channel patch–clamp, under cell-attached or -detached conditions as described in detail previously (35). In brief, cells were placed in an open, constantly perfused chamber (37°C). Solutions were designed to measure chloride currents only. In the cell-attached mode, bath and pipette were filled with a solution containing: 145 mM N-methyl-d-glucamine chloride (NMDGCl), 1.7 mM CaCl2, 1 mM MgCl, 10 mM Hepes, and 10 mM glucose ( pH 7.3). Single-channel conductance was determined by clamping voltage from −80 to 80 mV in 20-mV steps. For calculation of open probabilities, the apparent number of channels per patch was estimated by dividing the maximal current with the single-channel current. In the whole cell mode, pipette solution contained 145 mM NMDGCl, 0.1 mM EGTA, 1 mM MgCl2, 10 mM Hepes, 10 mM glucose, and 5 mM MgATP (pH 7.3). For continuous whole cell recordings, membrane potential was clamped to −60 mV. Current–voltage relations were generated from superimposed 200-ms voltage steps from −20 to 100 mV in 20-mV steps. Cells were stimulated with 10 μM forskolin (Calbiochem) in the bath solution. Subunit of catalytic protein kinase A (Promega) was used at 50 units/ml.
RESULTS
Construction of Functional Short Forms of CFTR.
Because of the limitations of the AAV packaging size, it is difficult to package a CFTR gene with an efficient promoter into an AAV vector. To achieve optimal levels of CFTR expression, we tested the possibility of reducing the size of CFTR by introducing deletions within the coding sequence, as well as by removing the untranslated regions. This approach was suggested by the results of structural–functional analyses of the CFTR gene showing that certain regions of the CFTR are not essential for the function (36, 37).
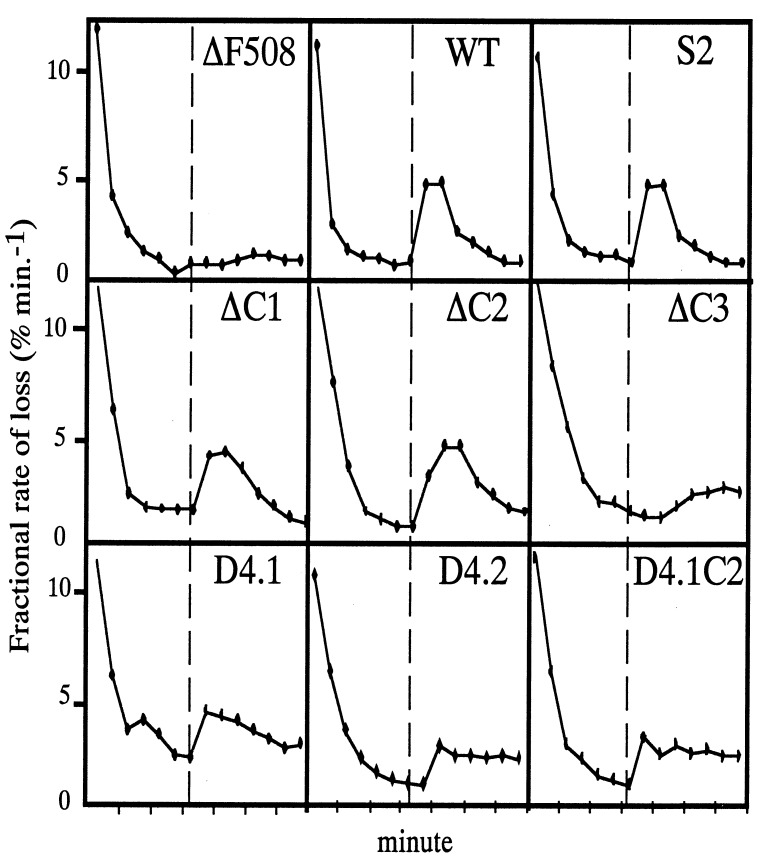
To reduce the size of the coding region while maintaining the protein function, we conducted deletion analysis of the CFTR gene. The full length CFTR cDNA encodes a protein that is predicted to have a N-terminal loop region, 12 transmembrane domains, two nucleotide binding domains, a regulatory (R) domain, and a cytoplasmic tail region (1, 2). Our strategy was to delete the N- and C-terminal sequences and the regions with few or no naturally occurring missense mutations. The lack of missense mutations in those regions suggests that these mutations may not affect the function of the CFTR protein and therefore are not detected by genetic screening of CF patients. We have made 20 deletions ranging from 51 to 360 bp at the N-terminal, cytoplasmic tail region, transmembrane domains, the loop regions between transmembrane domains, and the R domain. The regions where we have introduced deletions are diagrammed schematically in Fig. 1. The function of these modified CFTR genes was analyzed by using I125 efflux of cells stably transfected with each CFTR gene containing deletions (Fig. 2). We identified two nonessential regions by using this deletional screening approach. The first region is the cytoplasmic domain between nucleotides 4,441 and 4,572 (C-deletion) as defined by three deletional mutants, ΔC1, ΔC2, and ΔC3, which contain progressive deletions from the C terminus of the CFTR protein (see Table 1 for details). ΔC1 and ΔC2, which removed 23 and 44 aa residues, did not affect CFTR function as determined by the 125I efflux assays. The second region is a stretch of sequence between nucleotide 2,521 and 2,646 (R-deletion) that encodes the flexible region of the R domain. This region also was defined by three deletion mutants, D4.1, D4.2, and D4.3, which contain deletions progressively extending from the flexible region into the globular region of the R domain. D4.1, which removed 40 aa residues demonstrated a wild type-like Cl− channel activity. D4.2, with a 80-aa deletion, showed a delayed activation response to forskolin stimulation, and D4.3, with a 120-aa deletion that extended into the globular region of the R-domain, did not show a significant amount Cl− channel activity. Combining the C deletion (ΔC2) and R deletion (D4.1) resulted in a cDNA (D4.1C2) encoding an even smaller CFTR protein with wild-type CFTR activity.
Figure 1.
Locations of deletions made in the CFTR gene; 16 of the 20 deletions are shown. The CFTR gene is represented by patterned boxes. The gray regions represent loop regions, solid black boxes indicate transmembrane regions, and the dotted oval represents the globular portion of the R-domain. The two nucleotide binding domains are represented by dashed boxes. Deletions are represented by horizontal bars. Black bars represent internal deletions. Gray bars and hatched bars represents N- and C-terminal truncations, respectively. The numbers indicate the sizes of the deletion in nucleotides. Deletions that retain CFTR function are enclosed.
Figure 2.
Iodine efflux assays of CFTR constructs stably transfected into human 293 cells. Samples were taken at 1-min intervals. Forskolin was added at the dashed vertical line. The slope of the initial increase in rate of loss was taken as an index of CFTR activity. The ΔF508 is a negative control. WT is the 4.7-kb wild-type CFTR cDNA, and S2 is the cDNA containing the entire coding sequence but with only a 30-bp untranslated region at the 5′-end.
Comparison of Promoter Strengths in AAV Vectors.
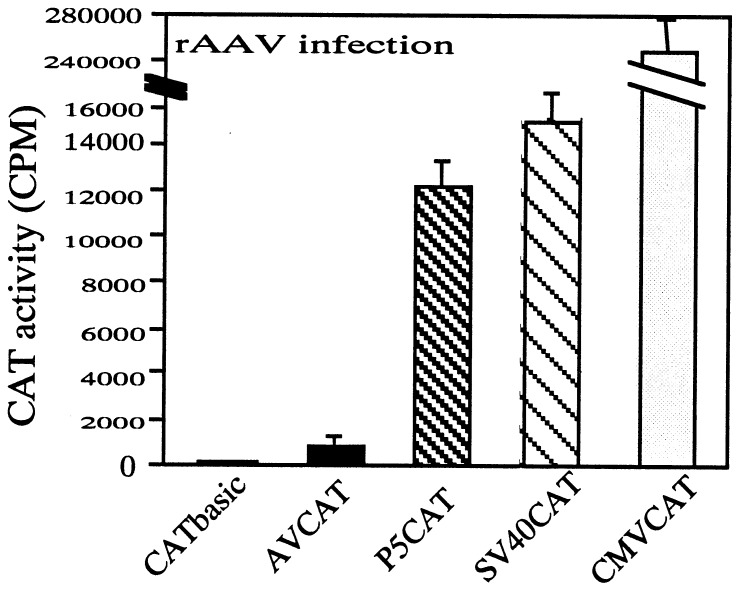
The second prerequisite for efficient expression of CFTR from an AAV vector is an efficient promoter. With deletions in the CFTR gene that we have made, it becomes possible to incorporate a promoter sequence of up to 500 bp in length. However, a shorter promoter is more desirable because the packaging of AAV favors vectors smaller than the size limit. In addition, the vector also must have room for a minimal polyadenylation signal (65 bp) needed for efficient gene expression. To search for the optimal promoter for expression of the CFTR gene, we analyzed several truncated viral promoters by reducing the flanking sequences and compared their activity with the CMVie promoter. The tested promoters were SV40e and the AAV P5 promoter. We also included a construct for which only the ITR sequence was used as the promoter. The activities of the promoters were analyzed by using AAV-mediated gene transfer with the CAT gene as a reporter. The AAV context is important; early experiments have suggested that the ITR sequences of AAV may influence the activity of adjacent promoters, presumably because of its secondary structures and regulatory elements for the P5 promoter. The transcriptional activities of these truncated promoters are shown in Fig. 3. The level of CAT expression from the ITR sequence was only a few-fold over the background expression from a plasmid carrying a CAT gene without any promoter. Both AAV P5 promoter (150 bp) and SV40e promoter (200 bp) transcribed CAT gene at levels that were 120–160 times higher than the background level. Although the CMVie promoter showed 15–20 times higher activity than P5 and SV40 promoters, the size of CMVie promoter (650 bp) would prevent its efficient packaging even with the shortened CFTR gene into an AAV vector.
Figure 3.
Comparison of promoter strengths. The promoter activities are represented by CAT activities indicated on the vertical axis. Each promoter is indicated on the horizontal axis. CATBasic is a CAT gene containing plasmid without a promoter. In AVCAT, the CAT gene is expressed from the AAV ITR. P5CAT contains the AAV ITR and P5 promoter. SVCAT and CMVCAT express the CAT gene from SV40e and CMVie promoter, respectively.
Efficient Transfer of Shortened CFTR Gene with AAV Vectors (Fig. 4).
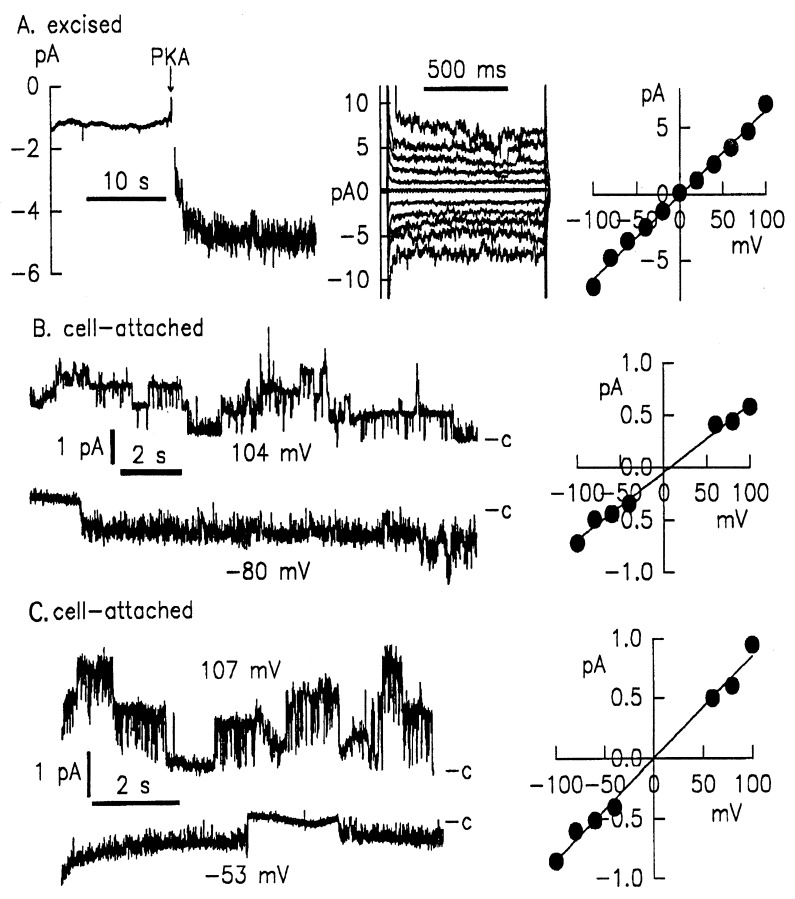
Figure 4.
Patch–clamp studies of HeLa cells infected with rAAV vectors containing functional CFTR cDNAs. (Left, A) Excised membrane patch of cells infected with AAVP5ΔC2. PKA treatment readily activated a current carried by multiple channels in this patch. I/V protocol applied during PKA treatment (middle graph) and I/V relations (right graph) show that the current is time- and voltage-independent. The total activated conductance in this patch was 64pS, equivalent to 16–20 CFTR channels. (B and C) Cell-attached, single-channel patch–clamp recording on HeLa cells infected with AAVp5ΔC2 or AAVp5D4.1C2, respectively. Cells were stimulated with forskolin and clamped to potentials as indicated. In both B and C, top and bottom curves show recordings containing at least three channels. Right graphs show linear current–voltage relations yielding a single channel conductance of 6.1pS + 0.27 (n = 6) for AAVp5ΔC2 or 8.7 pS + 0.16 (n = 6) for AAVP5D4.1C2. Whole-cell patch–clamp analysis of AAV–CFTR transduced cells.
To evaluate the AAV-mediated transduction of the short CFTR genes, we analyzed CFTR function with patch–clamp techniques. We infected HeLa cells that do not express CFTR with equal volumes of viral lysate containing AAVp5ΔC2 or AAVp5D4.1C2 (an AAV vector containing ΔC2 or D4.1C2 driven by a P5 promoter) and analyzed CFTR function with cell-attached and excised patch–clamp recordings. Fig. 4A shows the excised patch–clamp recording of cells transduced with AAVp5ΔC2. Treatment with protein kinase (PKA) readily activated current carried by multiple channels present in a single patch (Fig. 4A, left graph). Current–voltage (I/V) protocol applied during PKA treatment (Fig. 4A, middle graph) and I/V relations (Fig. 4A, right graph) show that the current in the excised mode was time- and voltage-independent. The total activated conductance in the patch was 64pS, which is equivalent to ≈16–20 Cl channels (assuming an open probability of 0.5 and 6–8pS for single CFTR). This indicated high levels of CFTR expression from the AAV vector.
With AAV-mediated transduction, we also were able to investigate the single-channel properties of the shortened CFTR gene (Fig. 4, B and C). In cells infected with AAVp5ΔC2, cell-attached recordings showed a linear Cl− channel with a conductance of 6.4 ± 0.16 pS (n = 5) (Fig. 4B, left graph). Patch–clamp recording of cells infected with AAVp5D4.1C2 (Fig. 4C, left graph) showed a significantly larger Cl− channel with a conductance of 8.7 ± 0.6 pS (n = 6). I/V relation curves (Fig. 4 B and C, right graphs) showed both channels to be time- and voltage-independent. Of interest, both ΔC2 and D4.1C2 exhibited very long open bursts and long closed times. During the stimulation, the average open probability was >0.5, comparable to the wild-type CFTR. These recordings confirmed that the CFTR deletion mutants form functional and cAMP/PKA-regulated Cl channels. Their biophysical characteristics (i.e., time -and voltage-independent activation and open probability) were not affected by the deletions, and their conductance was in the reported range of wild-type CFTR. The notable difference of the shortened CFTR compared with wtCFTR was the longer open and closed times.
Shorter CFTR Genes Are More Efficient in Transferring CFTR Function into Target Cells (Fig. 5).
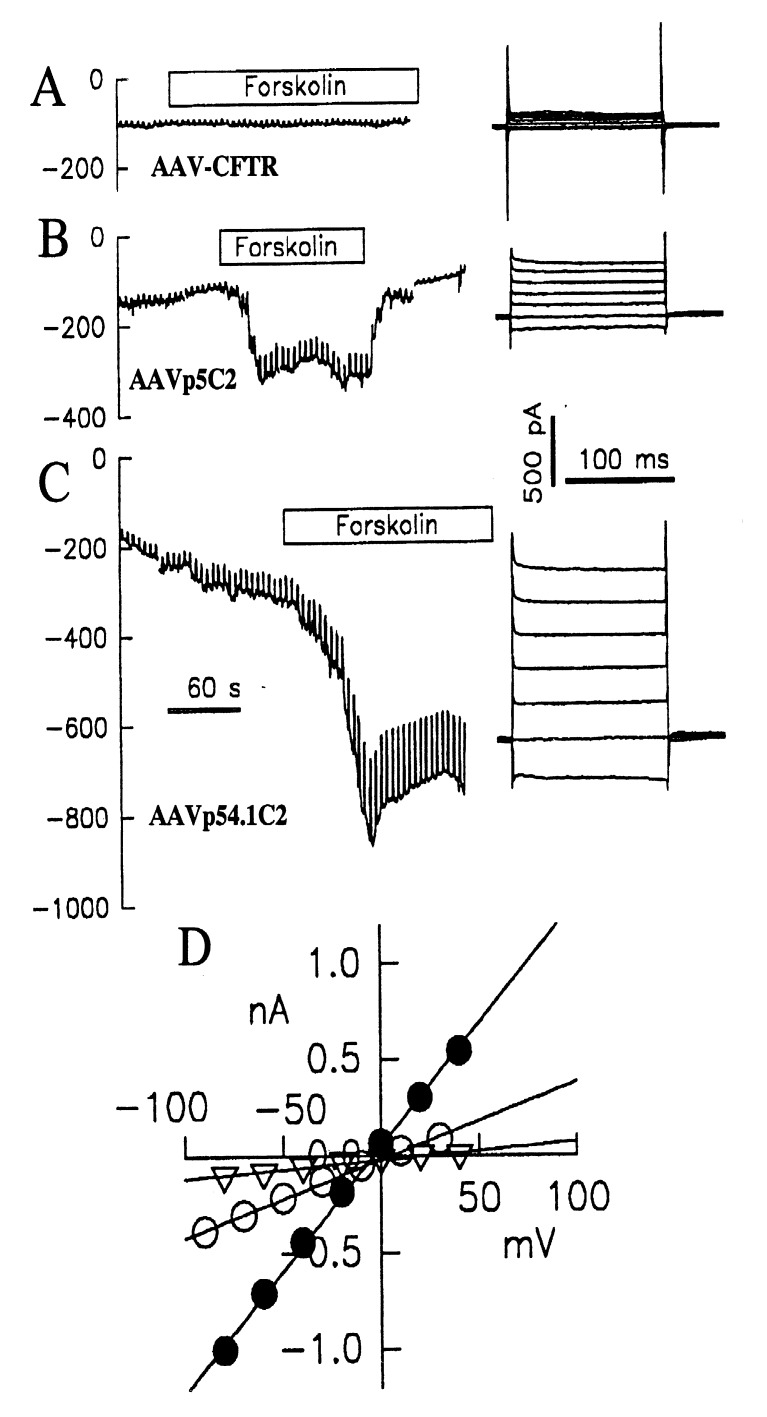
Figure 5.
Patch–clamp recording of cells transduced with (A) AAV-CFTR in which a full length CFTR is driven by ITR, (B) AAVp5ΔC2, and (C) AAVp5D4.1C2. The durations of forskolin treatment are indicated. Downward curves indicate forskolin-activated Cl− current. (D) Whole-cell current–voltage relations of each CFTR Cl− channel, triangle: ITR–CFTR; open circle: AAVp5ΔC2; filled circle: AAVp5D4.1C2. Minimal conductance was detected in cells transduced with AAV–CFTR, and highest conduction was detected in cells transduced with AAVp5D4.1C2 as indicated by the slope of the I/V curve.
We further analyzed the efficiencies of the different AAV/CFTR vectors in transferring the CFTR gene into target cells. The levels of transferred CFTR function were assayed with whole-cell patch–clamp measurements. This technique is quantitative because it compares the levels of CFTR expression in cells by measuring the amount of the Cl− current conducted. In this experiment, cells were infected with equal amounts of AAV stocks produced under identical conditions. AAV-CFTR, the vector containing a full length CFTR gene controlled by the ITR, was included for comparison. Whole-cell patch–clamp recordings of cells transduced with AAV–CFTR (4,837 bp), AAVp5ΔC2 (4,853 bp), or AAVp54.1C2 (4727 bp) are shown (Fig. 5). A strong forskolin-induced Cl− current was detected in cells transduced with AAVp5ΔC2 (Fig. 5B) but not in cells transduced with AAV–CFTR that do not contain a heterologous promoter (Fig. 5) although they are similar in length. An even stronger forskolin-induced Cl− current was recorded in cells transduced with AAVp5D4.1C2 (Fig. 5C), indicating that more efficient transduction of CFTR gene by this shorter vector resulted in the expression of more CFTR channels. Both ΔC2 and D4.1C2 are driven by the same P5 promoter, so the higher level expression of AAVp5D4.1C2 is probably caused by the more efficient packaging into virions of this vector, which is shorter by 126 bp. This is consistent with our previous observations that packaging efficiency is affected greatly by vector size when it is close to the packaging limit (32). I/V protocol and current–voltage relation (Fig. 5D) showed the CFTR-typical time and voltage independence of currents typical of CFTR and greater conductance mediated by the shorter CFTR genes.
DISCUSSION
AAV vector has been shown to be effective in transferring the CFTR cDNA into airway epithelial cells both in vitro and in vivo. However, its utility as a vector for gene therapy of CF is limited by its inefficiency in expressing the CFTR mini-gene in target cells. This is because the CFTR gene has been packaged into the vector without an efficient promoter because of the constraint of the small packaging capacity of AAV. Our goal is to develop a strategy to circumvent the package limit and to achieve high levels of the CFTR gene expression from AAV vectors. In this study, we have demonstrated the feasibility of reducing the size of the CFTR gene by targeted deletions in the coding region and expressing CFTR gene at high levels by packaging efficient promoters with the shortened CFTR gene into AAV vectors.
Our approach is to identify nonessential sequences within the CFTR coding region. First, we considered the regions that might not be structurally critical to the CFTR function, such as the N terminus and C terminus, the flexible regions of the R domain, and the longer loop regions between the transmembrane domains. Second, we targeted the regions where few natural missense mutations have been found, suggesting that mutations in these regions might not affect the function of the CFTR. Third, we recreated some of the CFTR deletional mutants that were functional in early structure–function studies. By combining this information, we have designed 20 deletions, of which 3 were found to retain the CFTR function. By combining the two larger deletions, we obtained an even smaller functional CFTR. Patch-clamp analyses showed that deletions in these regions did not affect the characteristics, such as regulation, open probability, and time–voltage independence of the CFTR channel. In single-channel patch–clamp, we did notice that the sequence-deleted CFTRs have longer open bursts and that the CFTR containing double deletions formed a slightly larger channel (8.7pS) than the wild-type CFTR. However, these properties should not affect their ability to compensate for the CFTR defect, and the larger channel size might be beneficial because it would require fewer of the CFTR molecules to achieve the same levels of Cl− transfer.
One interesting observation was that deletion mutants similar to those that showed Cl− channel function in studies using oocytes (of African clawed frog) were not functional in our studies. This was still true even after we restored the signal peptide of CFTR that was deleted in early studies. This may be explained by the difference between amphibian oocytes and mammalian cells. For example, Δ508 mutant is fully functional in oocytes but not in human cells. Deletions in the R-domain have been suggested to cause increasing open probability of the Cl− channel or altered regulation (38). In our study, deletions beyond the flexible region that links the putative globular domain did not function. Early studies using a phage T-7/vaccinia expression system in HeLa cells also have suggested that the C terminus may contain nonessential sequence (36). Two of our C-terminal deletions (C1 deleted 23 aa and C2 deleted 44 aa) were also functional. However, a more extensive deletion (C3), which deleted a 72-aa residue resulted in a Cl− channel with lower and delayed activity. When 86 aa residues were deleted from the C terminus (C4, data not shown), the proteins were not functional. Therefore, C2 and C3 defined a boundary (amino acids 1,408–1,436) of essential sequences in the C-terminal region. This boundary can be narrowed further to 1,408–1,419 aa residues when considering the functional mutant that deleted 61 aa residues from the C terminus in an early study (36).
Shorter AAV vectors are more efficiently packaged. Although other deletions impaired the functional of CFTR protein in the current studies, it is still possible that smaller deletions in these and other regions may leave the CFTR functional. Even smaller functional CFTRs may be generated to allow incorporating larger and stronger promoters as well as tissue-specific promoters to express CFTR function from the AAV vectors.
With the shortened CFTR genes, we were able to incorporate efficient promoters into AAV vectors to increase the level of CFTR expression. The increase in CFTR transcription is more than 10 times over a vector in which the CFTR gene is driven by the ITR sequence of AAV. In addition, the smaller size of these vectors fosters efficient packaging into AAV virions, which will allow production of high titer vectors for gene therapy studies. These new AAV/CFTR vectors should improve the efficacy of AAV-mediated gene therapy of CF.
Acknowledgments
We thank Lisa Woldin for assistance in preparing this manuscript and Dieter Gruenert and Semyon Rubinchik for their critical reviews. This study was supported by grants from the American Cystic Fibrosis Foundation: CFF P895, Z160, DONG96P0, and DONG97P0 and by grants from the National Institutes of Health: DK/HL46177 and DK47766. Y.W.K. is an investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- CF
cystic fibrosis
- CFTR
CF transmembrane conductance regulator
- rAAV
recombinant adeno-associated virus
- ITR
inverted terminal repeat
- CAT
chloramphenicol acetyltransferase
References
- 1.Riordan J R, Rommens J M, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou J L, et al. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 2.Rommens J M, Iannuzzi M C, Kerem B, Drumm M L, Melmer G, Dean M, Rozmahel R, Cole J L, Kennedy D, Hidaka N, et al. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 3.Gregory R J, Cheng S H, Rich D P, Marshall J, Paul S, Hehir K, Ostedgaard L, Klinger K W, Welsh M J, Smith A E. Nature (London) 1990;347:382–386. doi: 10.1038/347382a0. [DOI] [PubMed] [Google Scholar]
- 4.Welsh M J, Anderson M P, Rich D P, Berger H A, Denning G M, Ostedgaard L S, Sheppard D N, Cheng S H, Gregory R J, Smith A E. Neuron. 1992;8:821–829. doi: 10.1016/0896-6273(92)90196-k. [DOI] [PubMed] [Google Scholar]
- 5.Frizzell R A, Rechkemmer G, Shoemaker R L. Science. 1986;233:558–560. doi: 10.1126/science.2425436. [DOI] [PubMed] [Google Scholar]
- 6.Li M, McCann J D, Liedtke C M, Nairn A C, Greengard P, Welsh M J. Nature (London) 1988;331:358–360. doi: 10.1038/331358a0. [DOI] [PubMed] [Google Scholar]
- 7.Welsh M J. FASEB J. 1990;4:2718–2725. doi: 10.1096/fasebj.4.10.1695593. [DOI] [PubMed] [Google Scholar]
- 8.Jiang C, Finkbeiner W E, Widdicombe J H, Miller S S. J Physiol (London) 1997;501:637–647. doi: 10.1111/j.1469-7793.1997.637bm.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith J J, Travis S M, Greenberg E P, Welsh M J. Cell. 1996;85:229–236. doi: 10.1016/s0092-8674(00)81099-5. [DOI] [PubMed] [Google Scholar]
- 10.Boat T F, Cheng P W. Acta Paediatr Scand Suppl. 1989;363:25–29. doi: 10.1111/apa.1989.78.s363.25. [DOI] [PubMed] [Google Scholar]
- 11.Collins F S. Science. 1992;256:774–779. doi: 10.1126/science.1375392. [DOI] [PubMed] [Google Scholar]
- 12.Drumm M L, Pope H A, Cliff W H, Rommens J M, Marvin S A, Tsui L C, Collins F S, Frizzell R A, Wilson J M. Cell. 1990;62:1227–1233. doi: 10.1016/0092-8674(90)90398-x. [DOI] [PubMed] [Google Scholar]
- 13.Rich D P, Anderson M P, Gregory R J, Cheng S H, Paul S, Jefferson D M, McCann J D, Klinger K W, Smith A E, Welsh M J. Nature (London) 1990;347:358–363. doi: 10.1038/347358a0. [DOI] [PubMed] [Google Scholar]
- 14.Rosenfeld M A, Chu C S, Seth P, Danel C, Banks T, Yoneyama K, Yoshimura K, Crystal R G. Hum Gene Ther. 1994;5:331–342. doi: 10.1089/hum.1994.5.3-331. [DOI] [PubMed] [Google Scholar]
- 15.Zabner J, Couture L A, Gregory R J, Graham S M, Smith A E, Welsh M J. Cell. 1993;75:207–216. doi: 10.1016/0092-8674(93)80063-k. [DOI] [PubMed] [Google Scholar]
- 16.Zabner J, Wadsworth S C, Smith A E, Welsh M J. Gene Ther. 1996;3:458–465. [PubMed] [Google Scholar]
- 17.Wilson J M, Engelhardt J F, Grossman M, Simon R H, Yang Y. Hum Gene Ther. 1994;5:501–519. doi: 10.1089/hum.1994.5.4-501. [DOI] [PubMed] [Google Scholar]
- 18.Engelhardt J F, Simon R H, Yang Y, Zepeda M, Weber P S, Doranz B, Grossman M, Wilson J M. Hum Gene Ther. 1993;4:759–769. doi: 10.1089/hum.1993.4.6-759. [DOI] [PubMed] [Google Scholar]
- 19.Crystal R G, Hirschowitz E, Lieberman M, Daly J, Kazam E, Henschke C, Yankelevitz D, Kemeny N, Silverstein R, Ohwada A, et al. Hum Gene Ther. 1997;8:985–1001. doi: 10.1089/hum.1997.8.8-985. [DOI] [PubMed] [Google Scholar]
- 20.Wilmott R W, Amin R S, Perez C R, Wert S E, Keller G, Boivin G P, Hirsch R, De Inocencio J, Lu P, Reising S F, et al. Hum Gene Ther. 1996;7:301–318. doi: 10.1089/hum.1996.7.3-301. [DOI] [PubMed] [Google Scholar]
- 21.Brody S L, Metzger M, Danel C, Rosenfeld M A, Crystal R G. Hum Gene Ther. 1994;5:821–836. doi: 10.1089/hum.1994.5.7-821. [DOI] [PubMed] [Google Scholar]
- 22.Yei S, Mittereder N, Tang K, O’Sullivan C, Trapnell B C. Gene Ther. 1994;1:192–200. [PubMed] [Google Scholar]
- 23.Yang Y, Su Q, Wilson J M. J Virol. 1996;70:7209–7212. doi: 10.1128/jvi.70.10.7209-7212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao X, Li J, Samulski R J. J Virol. 1996;70:8098–8108. doi: 10.1128/jvi.70.11.8098-8108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher K J, Jooss K, Alston J, Yang Y, Haecker S E, High K, Pathak R, Raper S E, Wilson J M. Nat Med. 1997;3:306–312. doi: 10.1038/nm0397-306. [DOI] [PubMed] [Google Scholar]
- 26.Clark K R, Sferra T J, Johnson P R. Hum Gene Ther. 1997;8:659–669. doi: 10.1089/hum.1997.8.6-659. [DOI] [PubMed] [Google Scholar]
- 27.Rubenstein R C, McVeigh U, Flotte T R, Guggino W B, Zeitlin P L. Gene Ther. 1997;4:384–392. doi: 10.1038/sj.gt.3300417. [DOI] [PubMed] [Google Scholar]
- 28.Flotte T R, Afione S A, Conrad C, McGrath S A, Solow R, Oka H, Zeitlin P L, Guggino W B, Carter B J. Proc Natl Acad Sci USA. 1993;90:10613–10617. doi: 10.1073/pnas.90.22.10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flotte, T. (1997) Pediatr. Pulmonol. 14, Suppl., 83.
- 30.Wagner J A, Moran M L, Messner A H, Daifuku R, Kouyama K, Desch J K, Manley S, Norbash A, Kang S, Conrad C K, et al. Lancet. 1998;351:1702–1703. [Google Scholar]
- 31.Flotte T R, Afione S A, Solow R, Drumm M L, Markakis D, Guggino W B, Zeitlin P L, Carter B J. J Biol Chem. 1993;268:3781–3790. [PubMed] [Google Scholar]
- 32.Dong J Y, Fan P D, Frizzell R A. Hum Gene Ther. 1996;7:2101–2112. doi: 10.1089/hum.1996.7.17-2101. [DOI] [PubMed] [Google Scholar]
- 33.Fan P D, Dong J Y. Hum Gene Ther. 1997;8:87–98. doi: 10.1089/hum.1997.8.1-87. [DOI] [PubMed] [Google Scholar]
- 34.Ohrui T, Shen B Q, Mrsny R J, Widdicombe J H. J Appl Physiol. 1995;78:1197–1202. doi: 10.1152/jappl.1995.78.3.1197. [DOI] [PubMed] [Google Scholar]
- 35.Fischer H, Illek B, Machen T E. J Physiol. 1995;489:745–754. doi: 10.1113/jphysiol.1995.sp021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rich D P, Gregory R J, Cheng S H, Smith A E, Welsh M J. Receptors Channels. 1993;1:221–232. [PubMed] [Google Scholar]
- 37.Schwiebert E M, Morales M M, Devidas S, Egan M E, Guggino W B. Proc Natl Acad Sci USA. 1998;95:2674–2679. doi: 10.1073/pnas.95.5.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rich D P, Gregory R J, Anderson M P, Manavalan P, Smith A E, Welsh M J. Science. 1991;253:205–207. doi: 10.1126/science.1712985. [DOI] [PubMed] [Google Scholar]