Abstract
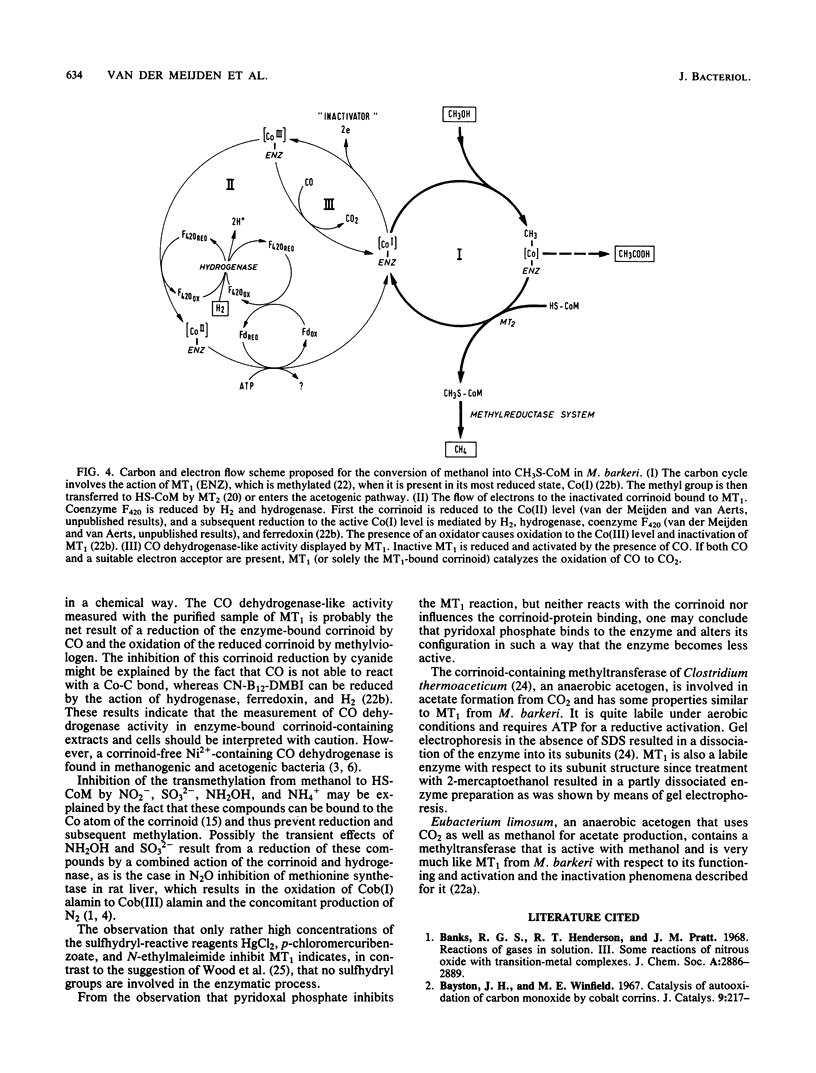
Methanol:5-hydroxybenzimidazolylcobamide methyltransferase from Methanosarcina barkeri has been purified to approximately 90% homogeneity by ion-exchange chromatography on DEAE-cellulose and QAE-A50 Sephadex columns. The molecular weight, estimated by gel electrophoresis, was found to be 122,000, and the enzyme contained two different subunits with molecular weights of 34,000 and 53,000, which indicates an alpha 2 beta structure. The enzyme contains three or four molecules of 5-hydroxybenzimidazolylcobamide, which could be removed by treatment of the enzyme with 2-mercaptoethanol or sodium dodecyl sulfate. In both cases the enzyme dissociated into its subunits. For stability, the enzyme required the presence of divalent cations such as Mg2+, Mn2+, Sr2+, Ca2+, or Ba2+. ATP, GTP, or CTP was needed in a reductive activation process of the enzyme. This activation was brought about by a mixture of H2, ferredoxin, and hydrogenase, but also by CO, which is thought to reduce the corrinoid chemically. The CO dehydrogenase-like activity of the methyltransferase is discussed.
Full text
PDF
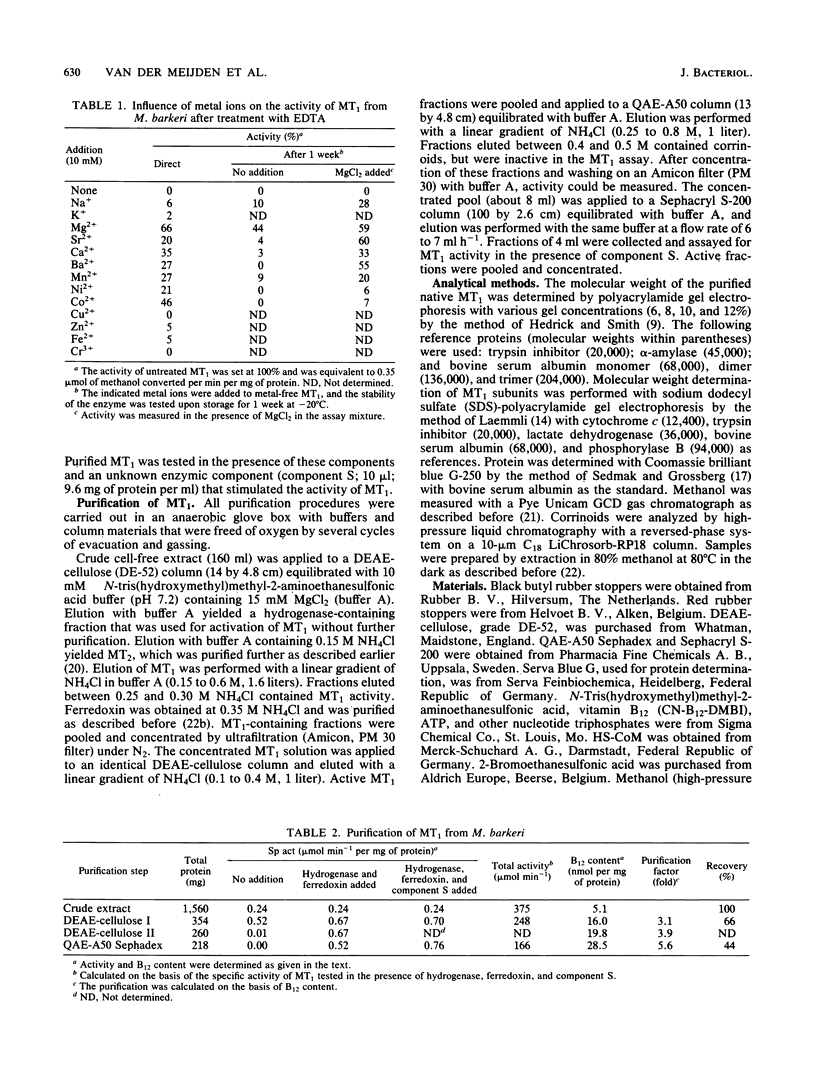
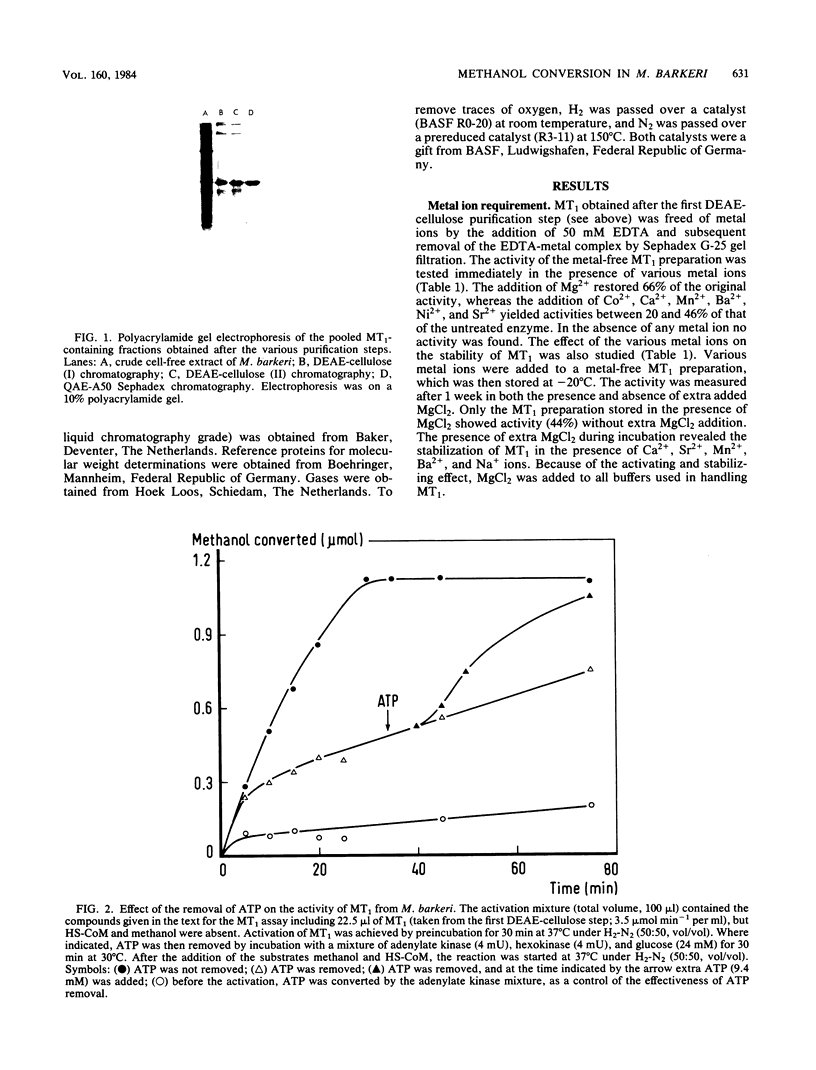
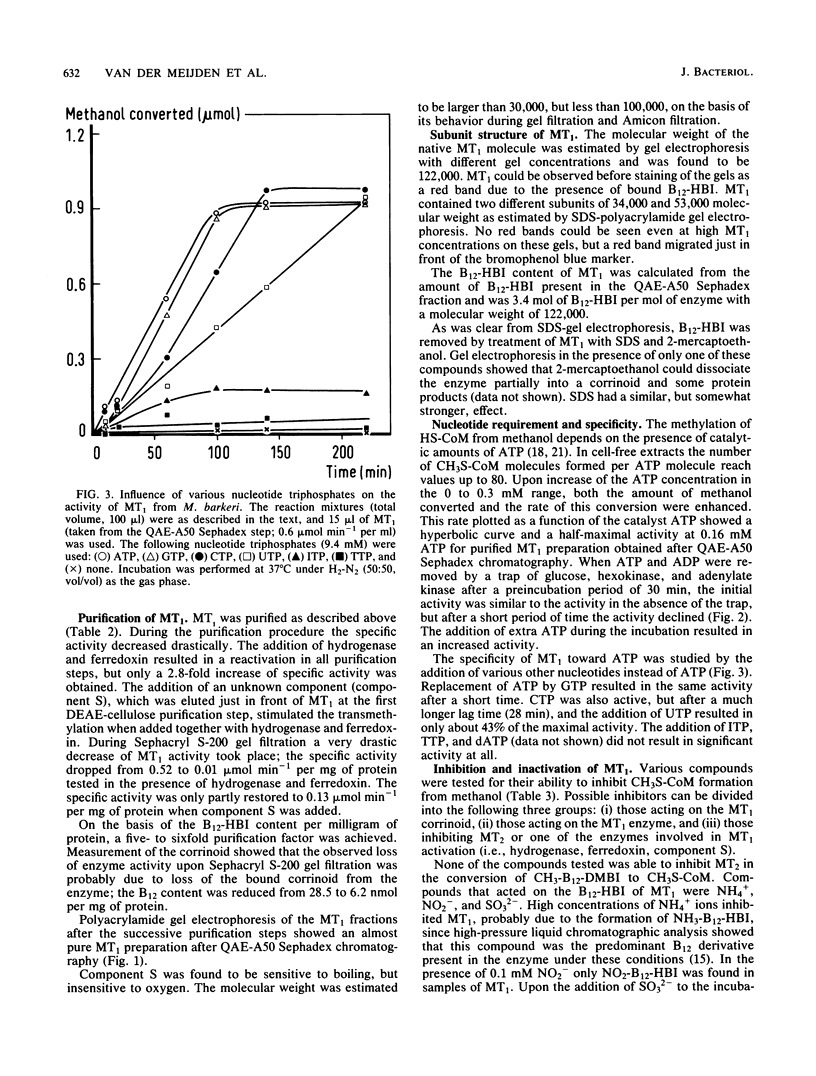


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Daniels L., Fuchs G., Thauer R. K., Zeikus J. G. Carbon monoxide oxidation by methanogenic bacteria. J Bacteriol. 1977 Oct;132(1):118–126. doi: 10.1128/jb.132.1.118-126.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon R., Lumb M., Perry J., Chanarin I., Minty B., Halsey M. J., Nunn J. F. Selective inactivation of vitamin B12 in rats by nitrous oxide. Lancet. 1978 Nov 11;2(8098):1023–1024. doi: 10.1016/s0140-6736(78)92341-3. [DOI] [PubMed] [Google Scholar]
- Diekert G. B., Thauer R. K. Carbon monoxide oxidation by Clostridium thermoaceticum and Clostridium formicoaceticum. J Bacteriol. 1978 Nov;136(2):597–606. doi: 10.1128/jb.136.2.597-606.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake H. L., Hu S. I., Wood H. G. Purification of carbon monoxide dehydrogenase, a nickel enzyme from Clostridium thermocaceticum. J Biol Chem. 1980 Aug 10;255(15):7174–7180. [PubMed] [Google Scholar]
- Ellefson W. L., Wolfe R. S. Role of component C in the methylreductase system of Methanobacterium. J Biol Chem. 1980 Sep 25;255(18):8388–8389. [PubMed] [Google Scholar]
- Gunsalus R. P., Wolfe R. S. ATP activation and properties of the methyl coenzyme M reductase system in Methanobacterium thermoautotrophicum. J Bacteriol. 1978 Sep;135(3):851–857. doi: 10.1128/jb.135.3.851-857.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick J. L., Smith A. J. Size and charge isomer separation and estimation of molecular weights of proteins by disc gel electrophoresis. Arch Biochem Biophys. 1968 Jul;126(1):155–164. doi: 10.1016/0003-9861(68)90569-9. [DOI] [PubMed] [Google Scholar]
- Hippe H., Caspari D., Fiebig K., Gottschalk G. Utilization of trimethylamine and other N-methyl compounds for growth and methane formation by Methanosarcina barkeri. Proc Natl Acad Sci U S A. 1979 Jan;76(1):494–498. doi: 10.1073/pnas.76.1.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten T. J., Bongaerts H. C., van der Drift C., Vogels G. D. Acetate, methanol and carbon dioxide as substrates for growth of Methanosarcina barkeri. Antonie Van Leeuwenhoek. 1980;46(6):601–610. doi: 10.1007/BF00394016. [DOI] [PubMed] [Google Scholar]
- Hutten T. J., De Jong M. H., Peeters B. P., van der Drift C., Vogels G. D. Coenzyme M derivatives and their effects on methane formation from carbon dioxide and methanol by cell extracts of Methanosarcina barkeri. J Bacteriol. 1981 Jan;145(1):27–34. doi: 10.1128/jb.145.1.27-34.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keltjens J. T., Whitman W. B., Caerteling C. G., van Kooten A. M., Wolfe R. S., Vogels G. D. Presence of coenzyme M derivatives in the prosthetic group (coenzyme MF430) of methylcoenzyme M reductase from Methanobacterium thermoautotrophicum. Biochem Biophys Res Commun. 1982 Sep 30;108(2):495–503. doi: 10.1016/0006-291x(82)90856-7. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Pol A., van der Drift C., Vogels G. D. Corrinoids from Methanosarcina barkeri: structure of the alpha-ligand. Biochem Biophys Res Commun. 1982 Sep 30;108(2):731–737. doi: 10.1016/0006-291x(82)90890-7. [DOI] [PubMed] [Google Scholar]
- Sedmak J. J., Grossberg S. E. A rapid, sensitive, and versatile assay for protein using Coomassie brilliant blue G250. Anal Biochem. 1977 May 1;79(1-2):544–552. doi: 10.1016/0003-2697(77)90428-6. [DOI] [PubMed] [Google Scholar]
- Shapiro S., Wolfe R. S. Methyl-coenzyme M, an intermediate in methanogenic dissimilation of C1 compounds by Methanosarcina barkeri. J Bacteriol. 1980 Feb;141(2):728–734. doi: 10.1128/jb.141.2.728-734.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor C. D., Wolfe R. S. A simplified assay for coenzyme M (HSCH2CH2SO3). Resolution of methylcobalamin-coenzyme M methyltransferase and use of sodium borohydride. J Biol Chem. 1974 Aug 10;249(15):4886–4890. [PubMed] [Google Scholar]
- Weimer P. J., Zeikus J. G. Acetate metabolism in Methanosarcina barkeri. Arch Microbiol. 1978 Nov 13;119(2):175–182. doi: 10.1007/BF00964270. [DOI] [PubMed] [Google Scholar]
- Welty F. K., Wood H. G. Purification of the "corrinoid" enzyme involved in the synthesis of acetate by Clostridium thermoaceticum. J Biol Chem. 1978 Aug 25;253(16):5832–5838. [PubMed] [Google Scholar]
- Wood J. M., Moura I., Moura J. J., Santos M. H., Xavier A. V., LeGall J., Scandellari M. Role of vitamin B12 in methyl transfer for methane biosynthesis by Methanosarcina barkeri. Science. 1982 Apr 16;216(4543):303–305. doi: 10.1126/science.7063887. [DOI] [PubMed] [Google Scholar]
- van der Meijden P., Heythuysen H. J., Pouwels A., Houwen F., van der Drift C., Vogels G. D. Methyltransferases involved in methanol conversion by Methanosarcina barkeri. Arch Microbiol. 1983 Jun;134(3):238–242. doi: 10.1007/BF00407765. [DOI] [PubMed] [Google Scholar]
- van der Meijden P., Heythuysen H. J., Sliepenbeek H. T., Houwen F. P., van der Drift C., Vogels G. D. Activation and inactivation of methanol: 2-mercaptoethanesulfonic acid methyltransferase from Methanosarcina barkeri. J Bacteriol. 1983 Jan;153(1):6–11. doi: 10.1128/jb.153.1.6-11.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meijden P., van der Lest C., van der Drift C., Vogels G. D. Reductive activation of methanol: 5-hydroxybenzimidazolylcobamide methyltransferase of Methanosarcina barkeri. Biochem Biophys Res Commun. 1984 Feb 14;118(3):760–766. doi: 10.1016/0006-291x(84)91460-8. [DOI] [PubMed] [Google Scholar]