Abstract
Linkage disequilibrium can be used for identifying associations between traits of interest and genetic markers. This study used mapped diversity array technology (DArT) markers to find associations with resistance to stem rust, leaf rust, yellow rust, and powdery mildew, plus grain yield in five historical wheat international multienvironment trials from the International Maize and Wheat Improvement Center (CIMMYT). Two linear mixed models were used to assess marker–trait associations incorporating information on population structure and covariance between relatives. An integrated map containing 813 DArT markers and 831 other markers was constructed. Several linkage disequilibrium clusters bearing multiple host plant resistance genes were found. Most of the associated markers were found in genomic regions where previous reports had found genes or quantitative trait loci (QTL) influencing the same traits, providing an independent validation of this approach. In addition, many new chromosome regions for disease resistance and grain yield were identified in the wheat genome. Phenotyping across up to 60 environments and years allowed modeling of genotype × environment interaction, thereby making possible the identification of markers contributing to both additive and additive × additive interaction effects of traits.
A useful new tool for crop genetic improvement is the identification of polymorphic markers associated with phenotypic variation for important traits by means of linkage disequilibrium (LD) between loci (Thornsberry et al. 2001; Flint-Garcia et al. 2003). A major advantage of this approach over conventional linkage mapping is that it does not require the time-consuming and expensive generation of specific genetic populations. LD is determined by the physical distance of the loci across chromosomes and has proven useful for dissecting complex traits because it offers fine-scale mapping due to the inclusion of historical recombination (Lynch and Walsh 1998). However, false positive correlation between markers and traits can arise in the absence of physical proximity due to population structure caused by admixture, mating system, and genetic drift or by artificial or natural selection during evolution, domestication, or plant improvement (Jannink and Walsh 2002). False associations can also be caused by alleles occurring at very low frequencies in the initial population (Breseghello and Sorrells 2006a,b). These factors create LD between loci that are not physically linked and cause a high rate of false positives when relating polymorphic markers to phenotypic trait variation. Thus, separating LD due to physical linkage from LD due to population structure is a critical prerequisite in association analyses.
Population structure can be quantified using Bayesian analysis, which has been effective for assigning individuals to subpopulations (Q matrix) using unlinked markers (Pritchard et al. 2000). Other multivariate statistical analyses such as classification (clustering) and ordination (scaling) can also be used to account for population structure (Kraakman et al. 2004). Population structure in modern breeding populations can be caused by complex pedigrees derived from crosses of parents with different levels of relatedness. Relationships between individuals can be detected by means of (1) marker-based estimation of the probability of identity by descent between individuals, (2) coefficient of parentage (COP) that measures the covariance between related individuals in a population (Parisseaux and Bernardo 2004), and (3) both types of analysis simultaneously (Arbelbide and Bernardo 2006; Malosetti et al. 2007). Yu et al. (2006) incorporated the outcome of population structure (Q matrix), with the estimation of relatedness between individuals obtained through the marker-based kinship matrix (K), into a unified linear mixed-model approach. Yu et al. (2006) showed that this approach effectively decreases type I error rates (false positives) and increases the power of the marker–trait association tests.
Successful application of association analysis requires comprehensive phenotypic data for modeling genotype × environment interaction (GE). However, an advantage of association analysis is that it can be based on historical phenotypic data from breeding trials, which in the case of the International Maize and Wheat Improvement Center (CIMMYT)'s wheat breeding programs is in plentiful supply. Linear mixed-model methodology applied to plant phenotypic data allows accurate prediction of genotypic performance using covariance structures that consider genetic associations between relatives included in the experiment. Using linear mixed-model methodology, a genetic covariance matrix can be estimated, and best linear unbiased predictors (BLUPs) can be obtained. BLUPs for simultaneously modeling GE and incorporating information on additive genetic variation have been previously suggested by Crossa et al. (2006) and Burgueño et al. (2007).
The CIMMYT wheat breeding program concentrates on producing stable high-yielding and widely adapted advanced breeding lines (Braun et al. 1996; Rajaram et al. 1996). CIMMYT bread wheat (Triticum aestivum L.) germplasm is known to contain various photoperiod insensitivity genes, dwarfing genes, and rust resistance genes (Trethowan et al. 2007). Grain yield and resistance to diseases, particularly to the rusts, are primary selection criteria, and the generation of closely linked molecular markers would speed up the breeding progress. CIMMYT's wheat breeding program annually distributes improved germplasm to a network of wheat research and breeding cooperators across the world who evaluate this material in many different environments and agronomic conditions. Phenotypic data from these trials have been cataloged, analyzed, and made available to the network. The data are used at CIMMYT to identify parents for future crosses and to drive the strategic incorporation of new genetic variability into advanced lines that are consequently able to cope with the dynamics of abiotic and biotic stresses.
In this study, we investigate the association of 242 diversity array technology (DArT) markers with resistance to stem (black) rust (caused by Puccinia graminis f.sp. tritici), leaf (brown) rust (caused by P. triticina), yellow (stripe) rust (caused by P. striiformis), powdery mildew (caused by Blumeria graminis f.sp. tritici), and grain yield in five historical CIMMYT elite spring wheat yield trials (ESWYT) conducted from 1979 to 2004 with data collected from up to 60 international environments. Two linear mixed models were used to assess marker–trait associations incorporating information on population structure and additive genetic covariance between relatives.
MATERIALS AND METHODS
Association populations and phenotypic data:
A total of 170 wheat lines derived from five CIMMYT elite spring wheat yield trials (ESWYT 1, ESWYT 6, ESWYT 10, ESWYT 20, and ESWYT 24) from 1979, 1984, 1988, 1999, and 2004, respectively, were included in this study. The data in each ESWYT were usually balanced, although for some traits data were recorded only from one replicate in some locations. There were no lines in common among the ESWYTs selected for this study, except for Genaro T81, which was included in ESWYTs 1 and 6. Depending on the trait and the specific ESWYT, the number of sites used for the various statistical analyses ranged from 6 to 60. Five traits were considered in this study: response to stem rust (SR) (percentage of infection), leaf rust (LR) (percentage of infection), yellow rust (YR) (percentage of infection), and powdery mildew (PM) (scored on a scale of 1–9) and grain yield (GY in tons per hectare). Association analyses were first performed for each of the five ESWYTs individually, and then the five ESWYTs were grouped into two sets related by year. ESWYTs 1, 6, and 10, which included 76 lines, formed ESWYT set 1, and ESWYTs 20 and 24, with 94 lines, formed ESWYT set 2. Unadjusted means of the five traits for each of the 170 wheat lines, their selection identifiers (SID), cross identifier (CID), line names, and ESWYT number are given in the additional information in supplemental Table S1 at http://www.genetics.org/supplemental/.
Depending on the trait and set, different sites, countries, and site–country combinations were used for the analyses. All five traits were recorded in all five ESWYTs in at least six sites, except SR, which was recorded only in sites of ESWYTs 1, 6, and 24.
DArT assay and map construction:
DArT markers were generated by Triticarte Pty. Ltd. (Canberra, Australia; http://www.triticarte.com.au), a whole-genome profiling service laboratory, as described by Akbari et al. (2006). The locus designations used by Triticarte Pty. Ltd. were adopted in this article, and DArT markers were named using the prefix “wPt,” followed by a unique numerical identifier. An integrated map of the DArT markers was developed using previous reported maps from diverse segregating populations, including: (1) Cranbrook × Halberd, 339 DArT (Akbari et al. 2006); (2) Arina × NK93604, 189 DArT (Semagn et al. 2006); (3) Avocet × Saar, 112 DArT (M. Lillemo, unpublished data); and (4) selected linkage groups of markers from nine different populations published by Triticarte Pty. Ltd. (http://www.triticarte.com.au). The integrated map was constructed with the software package CMTV (Sawkins et al. 2004); it included a total of 1644 markers, 813 of which were DArT markers and 831 of which were markers based on simple sequence repeats (SSRs), amplified fragment length polymorphisms (AFLPs), or restriction fragment length polymorphisms (RFLPs). Of a total of 813 DArT markers, 318 were polymorphic in the lines included in this study, and 242 of these were located on the final integrated map. A total of 76 polymorphic DArT markers could not be integrated into the final map (these are hereafter referred to as unmapped markers).
Population structure:
Possible population substructuring was investigated using the program STRUCTURE (Pritchard et al. 2000), on the basis of a set of 45 unlinked mapped DArT markers distributed evenly across the genome. Models with a putative number of subpopulations (K) from 1 to 20 without admixture and with noncorrelated allele frequencies were considered for each ESWYT. For the two ESWYT sets, K was set between 1 and 30. Lines within a subpopulation were assumed to be in Hardy–Weinberg equilibrium, as well as in linkage equilibrium (LE). In addition, the neighbor-joining clustering algorithm was run on the 170 lines (within ESWYTs and within combined ESWYT sets), using the Jaccard similarity coefficient as a proximity matrix with all 318 polymorphic DArT markers and the software DARWIN (Perrier et al. 2003). This step was undertaken to confirm the results of the population structure obtained from STRUCTURE.
Linkage disequilibrium:
Estimating LD between markers measures whether markers segregate independently or not. The program TASSEL (http://www.maizegenetics.net) was used to estimate the LD parameter r2 among loci, and the comparisonwise significance was computed with 1000 permutations. The r2 parameter was estimated for unlinked loci and for loci on the same chromosome. The latter was plotted for each ESWYT against genetic distance measured in centimorgans. A critical value for r2, as evidence of linkage, was derived using the 95% percentile of unlinked loci according to Breseghello and Sorrells (2006a). If, within a chromosome region, all pairs of adjacent loci were in LD, this region was referred to as an LD block (LDb).
The COP matrix:
The COP between individuals i and i′ is the probability that an allele from a randomly selected locus in individual i is identical by descent with an allele randomly selected from the same locus in individual i′ (Cockerham 1971). The genetic covariance between relatives due to their additive genetic effects is equal to two times the coefficient of parentage (Kempthorne 1969) (COP = fii′) times the additive genetic variance; i.e., 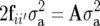 , where A is the additive relationship matrix and
, where A is the additive relationship matrix and 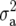 is the additive genetic variance. In the case of self-pollinating species with successive generations of inbreeding, COP between sister lines decreases with increasing numbers of generations following the split from the common ancestor. The Browse application of the International Crop Information System (ICIS), as described in http://cropwiki.irri.org/icis/index.php/TDM_GMS_Browse (McLaren et al. 2005), was used for deriving the relationship matrix; this application accounts for selection as well as inbreeding and improves the accuracy of breeding value estimation. The different ESWYTs had different numbers of sister line groups (ranging from 2 to 5). Matrix fii′ was computed for the 76 lines included in ESWYT set 1 and for the 96 lines of ESWYT set 2.
is the additive genetic variance. In the case of self-pollinating species with successive generations of inbreeding, COP between sister lines decreases with increasing numbers of generations following the split from the common ancestor. The Browse application of the International Crop Information System (ICIS), as described in http://cropwiki.irri.org/icis/index.php/TDM_GMS_Browse (McLaren et al. 2005), was used for deriving the relationship matrix; this application accounts for selection as well as inbreeding and improves the accuracy of breeding value estimation. The different ESWYTs had different numbers of sister line groups (ranging from 2 to 5). Matrix fii′ was computed for the 76 lines included in ESWYT set 1 and for the 96 lines of ESWYT set 2.
Linear mixed models used for association analyses:
The linear mixed model (LMM) was used to control the broad level of population structure inferred from Pritchard's STRUCTURE using the Q matrix and the level of relatedness between wheat lines using the relationship A matrix. Two kinds of linear mixed models (LMM1 and LMM2) were used to fit data from g lines, s sites, and r replicates (at each site) for modeling association of phenotypic traits with m markers. LMM1 is similar to some of the models proposed by Smith et al. (2002) for modeling GE and by Crossa et al. (2006) and Burgueño et al. (2007) for modeling GE with matrix A. In LMM2, markers are included as covariates for modeling the fixed effects of marker × environment interaction in a manner similar to the partition proposed in the factorial regression model (van Eeuwijk et al. 1996).
Linear mixed model 1 with covariance between relatives for modeling line and line × environment interaction:
Linear mixed model 1 (LMM1), which uses the relationship of the lines measured by the g × g A matrix for modeling GE, is
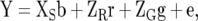 |
(1) |
where 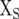 is the design matrix relating Y to the fixed effects of sites (b), and
is the design matrix relating Y to the fixed effects of sites (b), and 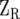 and
and 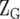 are the design matrices relating Y to the random effects of replicates within sites (r) and lines within sites (g), respectively. Vectors r, g, and e are assumed to be normally distributed, with zero mean vectors and variance–covariance (VCV) matrices R, G, and E, respectively. The VCV matrix G, which combines the main effect of lines and GE, can be represented as
are the design matrices relating Y to the random effects of replicates within sites (r) and lines within sites (g), respectively. Vectors r, g, and e are assumed to be normally distributed, with zero mean vectors and variance–covariance (VCV) matrices R, G, and E, respectively. The VCV matrix G, which combines the main effect of lines and GE, can be represented as 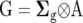 , where ⊗ is the Kronecker product operator, and the jth diagonal element of the
, where ⊗ is the Kronecker product operator, and the jth diagonal element of the 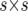 matrix
matrix 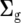 is the additive genetic variance
is the additive genetic variance 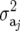 within the jth site, and the jjth element is the additive genetic covariance
within the jth site, and the jjth element is the additive genetic covariance 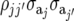 between sites j and j′; thus,
between sites j and j′; thus, 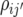 is the correlation of additive genetic effects between sites j and j′. The VCV matrix G was modeled using the factor analytical model (Smith et al. 2002; Crossa et al. 2006; Burgueño et al. 2007).
is the correlation of additive genetic effects between sites j and j′. The VCV matrix G was modeled using the factor analytical model (Smith et al. 2002; Crossa et al. 2006; Burgueño et al. 2007).
For each DArT marker, BLUPs of the lines obtained were used for contrasts of the predictable function of no difference between BLUPs with the mth DArT marker = 1 and BLUPs of lines with the mth DArT marker = 0. This was done using the VCV matrix of the BLUPs of lines obtained from LMM1. The contrasts between BLUPs were calculated by weighting each line with its probability of membership of each subpopulation obtained from matrix Q. The contrasts were performed for each marker at each subpopulation, as well as for each marker over all subpopulations.
A variant of LMM1 was proposed by Burgueño et al. (2007) for partitioning the total genetic effect (g) of a line into additive (a) and additive × additive (i) effects, as in Oakey et al. (2006). The covariance between individuals due to additive × additive effects in matrix notation is 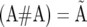 (where # is the elementwise multiplication operator). This model (with the partition of g into a and i) was used only for GY in each individual ESWYT and in ESWYT set 2. Thus, results from these trials were presented in terms of GY, GYA, and GYAA, which refer to the total genetic effect, the additive effects, and the additive × additive effects, respectively. Highly unbalanced data for other traits in ESWYT set 1 prevented fitting LMM1 with g = a + i.
(where # is the elementwise multiplication operator). This model (with the partition of g into a and i) was used only for GY in each individual ESWYT and in ESWYT set 2. Thus, results from these trials were presented in terms of GY, GYA, and GYAA, which refer to the total genetic effect, the additive effects, and the additive × additive effects, respectively. Highly unbalanced data for other traits in ESWYT set 1 prevented fitting LMM1 with g = a + i.
Linear mixed model 2 with covariance between relatives for modeling the polygenic effects of lines and including marker × environment interaction as fixed effects:
The best linear unbiased estimates (BLUEs) obtained from the individual-site analysis and their weights were used in the second-stage across-site analysis to calculate the BLUPs of the lines. The weights for the ith genotype in the jth location, wij, were computed as suggested by Cullis et al. (1996), 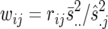 , where rij is the number of replicates for the ith genotype in the jth environment,
, where rij is the number of replicates for the ith genotype in the jth environment, 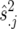 is the estimated error variance for the jth environment, and
is the estimated error variance for the jth environment, and 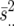 is the pool error variance. The second-stage across-site analysis uses linear mixed model 2 (LMM2) with random sites (s), random polygenic effects of lines (g), and fixed covariables from matrix Q of population structure and partitions the GE into marker × environment interactions as fixed covariable effects
is the pool error variance. The second-stage across-site analysis uses linear mixed model 2 (LMM2) with random sites (s), random polygenic effects of lines (g), and fixed covariables from matrix Q of population structure and partitions the GE into marker × environment interactions as fixed covariable effects
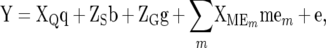 |
(2) |
where 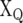 is the design matrix of the fixed effects of population structure (covariate) (q);
is the design matrix of the fixed effects of population structure (covariate) (q); 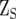 and
and 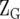 are the design matrices of the random effect of sites (s) and lines (g), and
are the design matrices of the random effect of sites (s) and lines (g), and 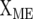 is the design matrix for the fixed effect of the marker × site interaction (me) for each of the m markers. The design matrix
is the design matrix for the fixed effect of the marker × site interaction (me) for each of the m markers. The design matrix 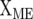 contains the allelic values of the markers, which for DArT markers are 0 and 1 (or −1 and 1). Vector g is assumed to be normally distributed with zero mean and VCV matrix
contains the allelic values of the markers, which for DArT markers are 0 and 1 (or −1 and 1). Vector g is assumed to be normally distributed with zero mean and VCV matrix 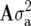 ; vectors of random effects of sites (b) and residual error (e) are assumed to have a simple VCV.
; vectors of random effects of sites (b) and residual error (e) are assumed to have a simple VCV.
Predictable functions of the contrast of the BLUPs of lines:
For the case of association analysis, for a given marker m, the null hypothesis of no difference between BLUPs of the lines with the mth DArT marker = 1 and BLUPs of the lines with the mth DArT marker = 0 was tested using a generalized t-statistic. The Wald z-statistics were also used for a simple contrast or Wald F-statistics for multiple contrasts. Asymptotically, the Wald statistics follow a chi-squared distribution.
Controlling the rate of false positives and the false discovery rate:
Critical P-values for assessing the significance of the null hypothesis were calculated using the Bonferroni genomewise error rate protection against rejecting a true null hypothesis (lower type I error rate). The testwise significance level was divided by the number of tests (markers). The control of the false discovery rate (FDR) for the test of each marker (Benjamini and Hochberg 1995) was used as an independent alternative method.
RESULTS
Variance component estimates are given in Table 1 for five traits in each individual ESWYT and in ESWYT sets 1 and 2. Site variability was important, but genetic as well as GE components were also of considerable size. The additive and additive × additive components and their interaction with sites for GY in ESWYTs 20 and 24, as well as in ESWYT set 2, were greater than those in ESWYTs 1, 6, and 10, probably due to the larger number of sites included in ESWYT set 2.
TABLE 1.
Estimates of environment (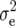 ), genetic (
), genetic (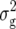 ), genetic × environment (
), genetic × environment (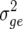 ), and pooled within-site error (
), and pooled within-site error ( ) variances from a complete random-effect model of five traits, grain yield (GY), leaf rust (LR), stem rust (SR), yellow (stripe) rust (YR), and powdery mildew (PM) for each of the ESWYTs and combined in ESWYT sets 1 and 2
) variances from a complete random-effect model of five traits, grain yield (GY), leaf rust (LR), stem rust (SR), yellow (stripe) rust (YR), and powdery mildew (PM) for each of the ESWYTs and combined in ESWYT sets 1 and 2
| Trait | 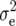 |
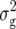 |
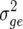 |
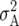 |
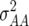 |
 |
|---|---|---|---|---|---|---|
| ESWYT 1 | ||||||
| GY | 3.757 | 0.0757 | 0.1982 | 0.033 | 0.0742 | 0.4663 |
| LR | 22.87 | 59.65 | 61.79 | — | — | 42.90 |
| PM | 2.473 | — | — | — | — | 1.092 |
| SR | 56.94 | 20.66 | 69.91 | — | — | 41.28 |
| YR | 57.40 | 0 | 230.8 | — | — | 61.53 |
| ESWYT 6 | ||||||
| GY | 3.717 | 0.0320 | 0.2383 | 0.0793 | 0.0583 | 0.5091 |
| LR | 90.98 | 8.826 | 235.4 | — | — | 65.14 |
| PM | 1.721 | 0.0052 | 1.399 | — | — | 1.257 |
| SR | 37.26 | 94.81 | 125.0 | — | — | 58.16 |
| YR | 89.67 | 65.31 | 267.3 | — | — | 64.31 |
| ESWYT 10 | ||||||
| GY | 5.948 | 0.0543 | 0.2761 | 0.0767 | 0.0841 | 0.3442 |
| LR | 206.4 | 0 | 187.8 | — | — | 10.95 |
| PM | 20.59 | 0.2527 | 1.363 | — | — | 0.1371 |
| SR | — | — | — | — | — | — |
| YR | 530.8 | 96.31 | 237.6 | — | — | 60.63 |
| Combined ESWYTs 1, 6, 10 (set 1) | ||||||
| GY | 0 | 0 | 1.171 | — | — | 0.6942 |
| LR | 54.89 | 23.13 | 140.3 | — | — | 98.16 |
| PM | 0 | 0.2090 | 1.296 | — | — | 0.8705 |
| SRa | 58.48 | 53.02 | 60.33 | — | — | 64.10 |
| YR | 203.7 | 281.9 | 232.0 | — | — | 59.21 |
| ESWYT 20 | ||||||
| GY | 4.713 | 0.0307 | 0.2879 | 0.1057 | 0.1062 | 0.5780 |
| LR | 103.1 | 37.26 | 85.64 | — | — | 87.08 |
| PM | 1.358 | 0.7604 | 0.9025 | — | — | 0.7412 |
| SR | — | — | — | — | — | — |
| YR | 26.20 | 177.0 | 114.8 | — | — | 94.57 |
| ESWYT 24 | ||||||
| GY | 5.303 | 0.0244 | 0.2592 | 0.2121 | 0.1680 | 0.4390 |
| LR | 374.6 | 11.23 | 26.73 | — | — | 89.15 |
| PM | 4.981 | 0.3801 | 1.069 | — | — | 1.775 |
| SR | 2.554 | 23.55 | 117.6 | — | — | 86.40 |
| YR | 55.28 | 137.3 | 200.8 | — | — | 75.59 |
| Combined ESWYTs 20 24 (set 2) | ||||||
| GY | 2.015 | 0.2968 | 0.1158 | 0.2943 | 0.2587 | 0.9439 |
| LR | 138.9 | 34.23 | 24.82 | — | — | 89.84 |
| PM | 0.3492 | 0.5968 | 0.9166 | — | — | 1.6256 |
| SRa | — | — | — | — | — | — |
| YR | 18.90 | 150.8 | 111.5 | — | — | 125.7 |
Estimates are shown of additive plus additive × environment (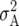 ), additive × additive plus additive × additive × environment (
), additive × additive plus additive × additive × environment (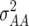 ) from LMM1 partitioning the total genotypic plus genotypic × environment effects (g) into additive plus additive × environment (a), and additive × additive plus additive × additive × environment (i) for GY (for ESWYT set 1 the LMM1 with g = a + i could not be fitted).
) from LMM1 partitioning the total genotypic plus genotypic × environment effects (g) into additive plus additive × environment (a), and additive × additive plus additive × additive × environment (i) for GY (for ESWYT set 1 the LMM1 with g = a + i could not be fitted).
Only ESWYTs 1 and 6 at set 1 and only ESWYT 24 at set 2.
Linkage disequilibrium:
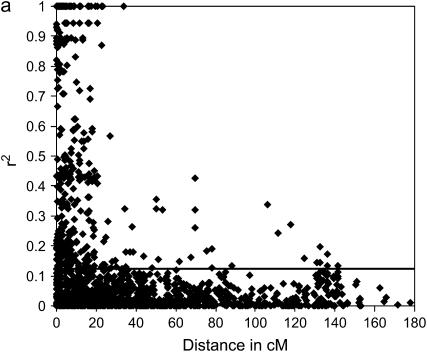
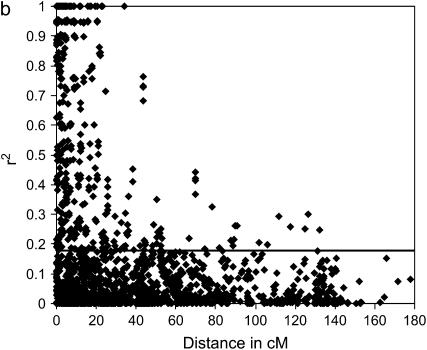
LD analyses were performed for each ESWYT individually, for ESWYT sets 1 and 2, and for both sets combined. The LD patterns were distinct for each individual ESWYT; however, for ESWYT set 1, ESWYT set 2, and the combined sets, the LD patterns were very similar. As LD breakdown depends on the number and relatedness of the lines under study, this result was not unexpected. The percentage of physically linked loci pairs located in significant LD blocks was 26.2% in ESWYT set 1 (r2 > 0.124, P < 0.01), 25.3% in ESWYT set 2 (r2 > 0.182, P < 0.01), and 27.6% in both sets combined (r2 > 0.115, P < 0.01). Pairwise r2 estimates were on average 0.169, 0.196, and 0.169 for ESWYT set 1, ESWYT set 2, and both sets combined, respectively. The percentages of unlinked pairs of loci that were in significant LD were 4.9% in ESWYT set 1, 5.0% in ESWYT set 2, and 5.0% in both sets combined. The estimated averages of r2 for unlinked loci were 0.03 in ESWYT set 1, 0.05 in ESWYT set 2, and 0.03 in both sets combined.
As depicted in Figure 1a for the LD of ESWYT set 1 and in Figure 1b for that of ESWYT set 2, LD decreases with increasing genetic map distance between marker loci. In all three cases, the r2 declines within ∼40 cM below critical values (indicating no linkage disequilibrium). This occurs at a larger distance than in previously reported studies of wheat (Breseghello and Sorrells 2006a), which focused on SSR markers across two chromosomes only. Since LD blocks in the seven chromosomal groups for each ESWYT set and in both sets combined were very similar, the LD blocks from both sets combined were adopted in the integrated map. A total of 43 LD blocks, with an average length of 9.93 cM, were detected across all chromosome groups. The longest LD blocks, with approximate lengths of 64.96 and 87.41 cM, respectively, were observed on chromosomes 1B and 4B.
Figure 1.—
Linkage disequilibrium (LD) (r2) as a function of map distance of 318 DArT markers in ESWYT set 1 (a) and ESWYT set 2 (b). Horizontal lines represent the critical value for LD significance.
Linkage maps:
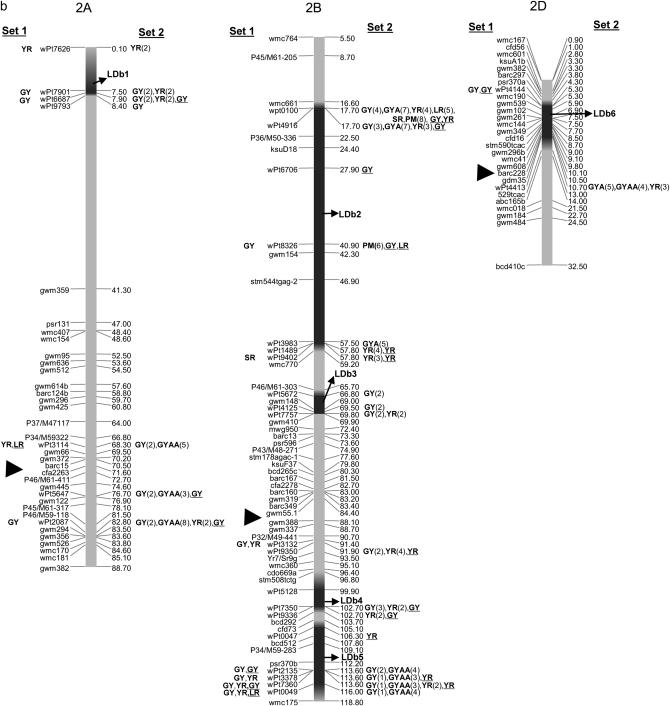
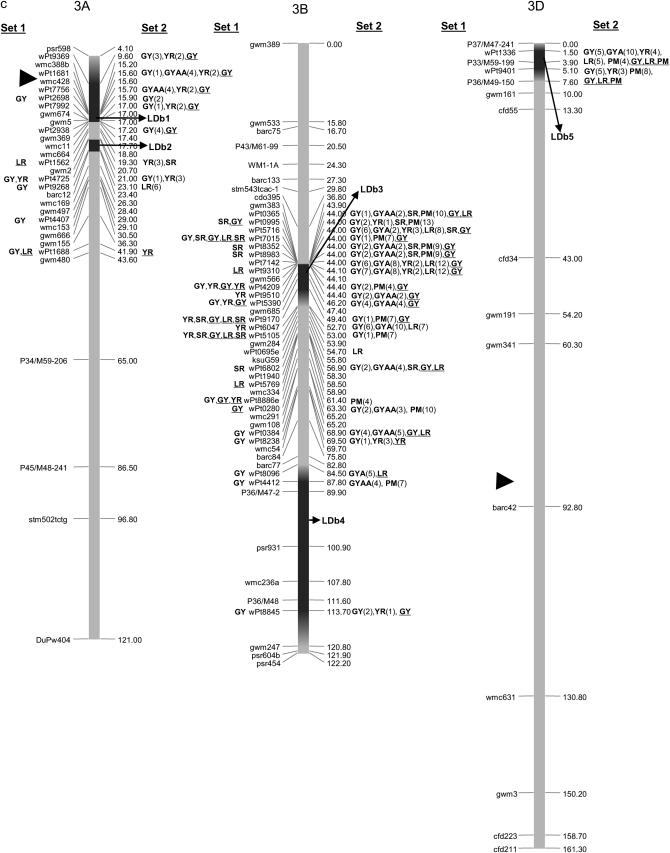
The integrated maps with 1644 markers (813 DArT's and 831 SSRs, AFLPs, or RFLPs) are displayed in supplemental Figures S1–S7 for chromosome groups 1–7, respectively, at http://www.genetics.org/supplemental/. Of the 813 DArT markers, only 318 were polymorphic in the 170 lines. Of these, 242 were located on the final integrated map, with 79, 149, and 20 DArT markers in genomes A, B, and D, respectively. No marker could be located on chromosome 6D due to low numbers of polymorphic DArT's per se and to insufficient anchor markers among the segregating populations utilized for map development. Polymorphic markers on the remaining chromosomes ranged from 2 DArT's on chromosome 5D to 20 DArT's on chromosome 7B. The power of testing marker–trait associations is reduced using markers with low frequencies; however, this was not a problem in this study, as the average frequency of DArT polymorphisms was 0.57. A frequency <5% for the absent score (“zero” allele) was observed for only one marker (wpt9124). For graphical simplicity we present the reduced maps in Figure 2, a–g, where a subset of the 831 SSR, AFLP, and RFLP markers and all 242 DArT markers corresponding to each chromosome group 1–7, respectively, are included.
Figure 2.—
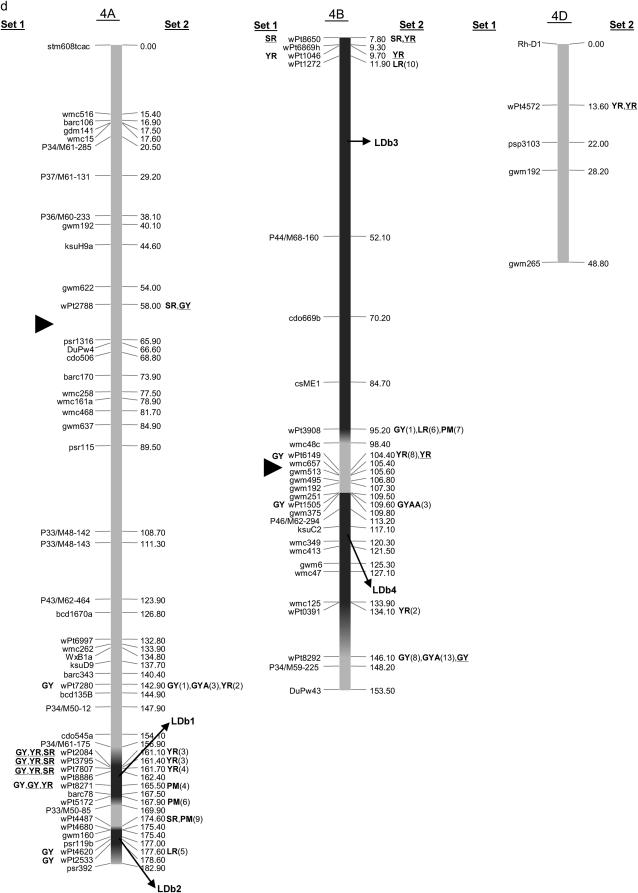
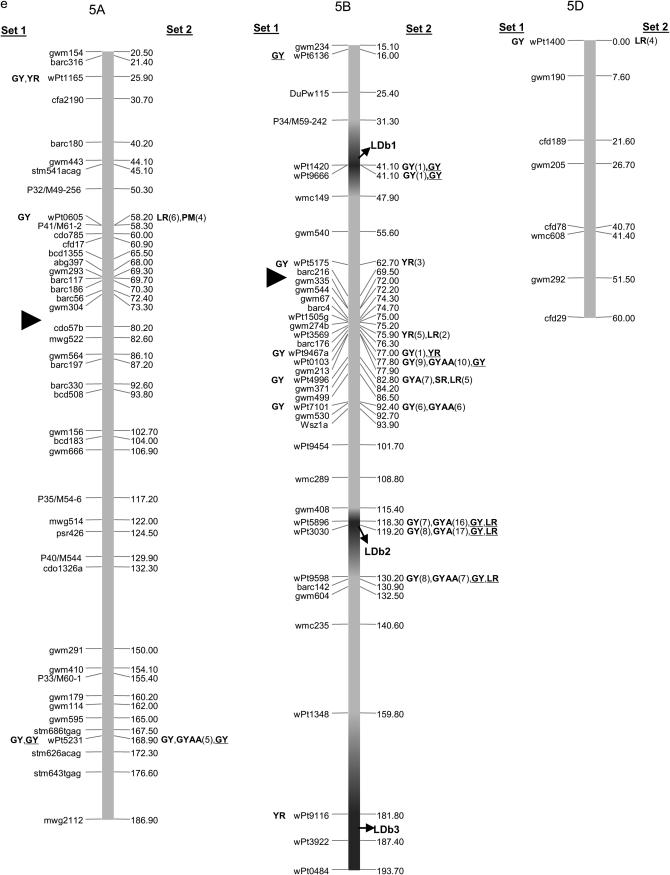
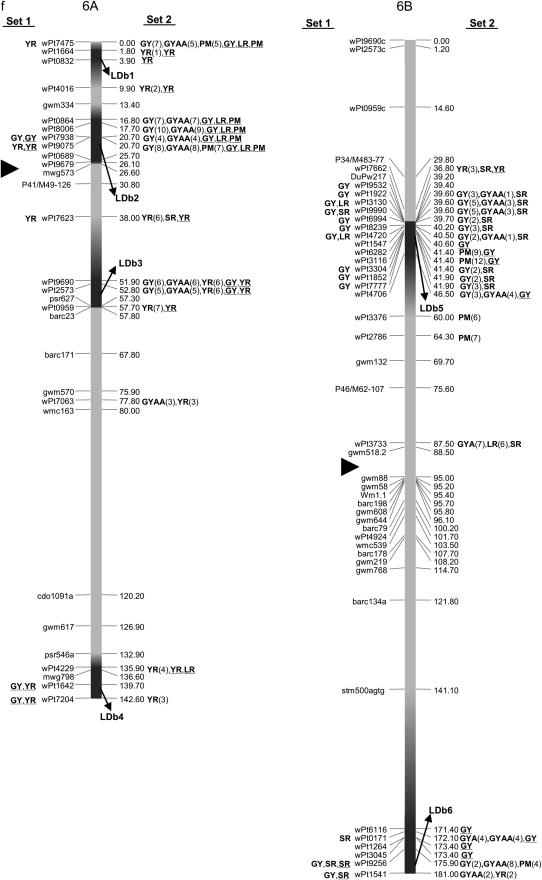
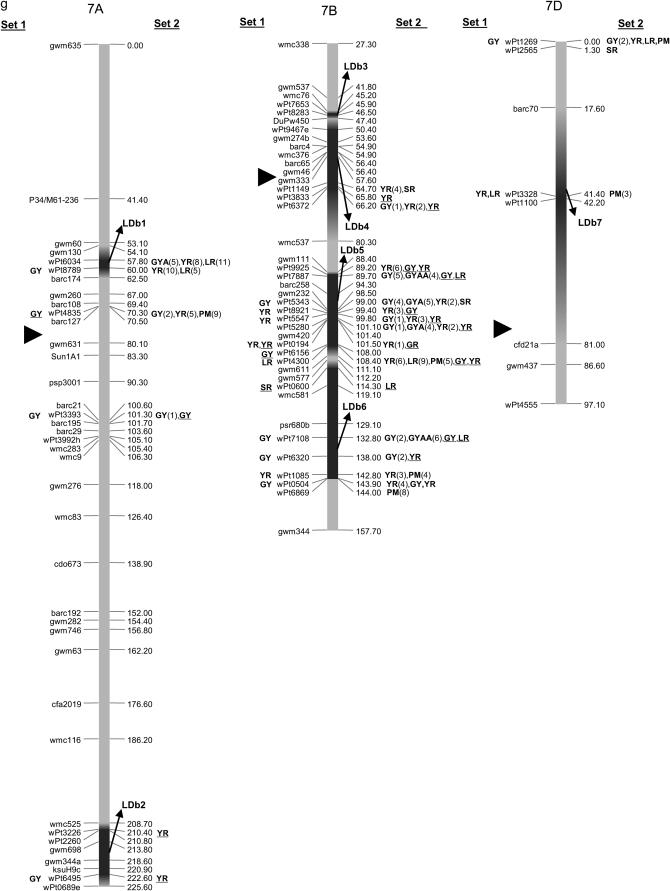
Map with reduced number of markers including the 242 DArT and other markers. Significant markers are shown for leaf rust (LR), stem rust (SR), yellow rust (YR), powdery mildew (PM), grain yield (GY), additive GY (GYA), and additive × additive GY (GYAA) for ESWYT set 1 (left side of the chromosome), and ESWYT set 2 (right side of the chromosome) for linear model 1 (not underlined) and linear model 2 (underlined). Numbers in parentheses refer to the variability explained by the marker as a percentage of the total variability for linear model 1. (a) Chromosome group 1 (1A, 1B, 1D). (b) Chromosome group 2 (2A, 2B, 2D). (c) Chromosome group 3 (3A, 3B, 3D). (d) Chromosome group 4 (4A, 4B, 4D). (e) Chromosome group 5 (5A, 5B, 5D). (f) Chromosome group 6 (6A, 6B). (g) Chromosome group 7 (7A, 7B, 7D). Approximate position of the centromere is marked with ▸.
Population structure:
Population structure was determined for each ESWYT individually. Model-based clustering analyses using the program STRUCTURE and the neighbor-joining clustering algorithm showed a pattern of substructure for ESWYT 20 only, which had seven subpopulations on the basis of the log-likelihood profile. Subsets of sister lines included in ESWYT 20 were assigned to the same subpopulation, except for the first subpopulation where three sister lines from different subsets were clustered. The STRUCTURE and neighbor-joining clustering algorithm analyses for the rest of the ESWYTs taken individually showed a pattern typical of unstructured populations. Taken as a set, the 76 lines contained in ESWYT set 1 were grouped into 16 subpopulations, while the 94 lines in ESWYT set 2 were grouped into 15 subpopulations (data not shown).
Association analyses of phenotypic traits:
Significant markers for each individual ESWYT and trait analyses are presented for LMM1 (supplemental Table S2 at http://www.genetics.org/supplemental/) and LMM2 (supplemental Table S3 at http://www.genetics.org/supplemental/) (the effect of each marker is given only for LMM1). The success in fitting the combined data for each ESWYT set depended on the trait, the number of common sites, countries, and site–country combinations and the LMM considered. For SR, results for ESWYT set 1 included ESWYTs 1 and 6, whereas results from set 2 included only ESWYT 24. For ESWYT set 1, LMM1 was fitted to GY but the model with the partition of g into a and i could not be fitted. For ESWYT set 1, PM was not included because no common sites or site–country combinations were found.
Several DArT markers on different chromosomes were significant for various traits simultaneously, and many unmapped DArT markers were significant as well. Only the results of association analysis for ESWYT sets 1 and 2 and for both LMMs are shown. To facilitate description and interpretation of the results, reference to numbered LDb's containing markers in LD is frequently made. The LDb's are highlighted in Figure 2, a–g.
Significant DArT markers for traits:
The mapped and unmapped DArT markers significantly associated with LR, SR, PM, YR, and GY (including GYA and GYAA) for ESWYT sets 1 and 2 and for LMM1 and LMM2 are summarized in Tables 2–6. In addition, we present the most recently published information on the chromosomal locations of genes and quantitative trait loci (QTL) for LR, SR, PM, and YR resistance and for GY, including genes for photoperiod insensitivity (Ppd), vernalization (Vrn), earliness per se (Eps), and dwarfism (Rht). Tables 2–6 summarize the information depicted in Figure 2, a–g. Significant DArT markers for YR and GY are widely spread across all chromosomes, with a total of 122 and 213 significantly associated DArT markers, respectively, vs. 87, 63, and 61 significant DArT markers for LR, SR, and PM, respectively. Chromosome groups 4 and 5 had the lowest number of significant DArT markers across all traits (47 and 35, respectively), whereas chromosome groups 1, 2, 3, 6, and 7 had a total of 132, 64, 99, 85, and 83 significant DArT markers, respectively. A comparison among the A, B, and D genomes is of less interest due to the drastically different numbers of DArT markers that were polymorphic within each genome.
TABLE 2.
Location of the significant DArT markers associated with leaf rust found in ESWYT set 1 (ESWYTs 1, 6, 10) and ESWYT set 2 (ESWYTs 20 and 24) using linear mixed model 1 (LMM1) and linear mixed model 2 (LMM2) for each chromosome and the reported Lr genes and QTLs
| Short arm (S)
|
Long arm (L)
|
Unknown arm
|
||||
|---|---|---|---|---|---|---|
| Chromosome | DArT (wPt) | Lr gene and QTL | DArT (wPt) | Lr gene and QTL | DArT (wPt) | Lr gene |
| 1A | 5374f, 2872, 4029, 6709 | Lr10 | 8016, 0128 | — | — | — |
| 1B | 1328, 3465, 4434, 0974, 6427, 8986, 1781, 5065, 2614, 5678, 5363, 6777, 5801, 6117, 6833, 8616, 2315 | Lr26, QTLab | 0944, 2526, 4129 | Lr46 | 2019, 5316, 1139 | Lr33, -44, -55, -51 |
| 1D | — | Lr21c QTLa | 3743 | — | — | Lr40, -42, -43 |
| 2A | 3114 | Lr17a, -17b,-35,-45 | — | Lr38 | 6207 | Lr11 |
| 2B | 0100, 8326 | Lr13, -16, -23 QTLdef | 0049 | Lr50 | 0094, 4559 | Lr35 |
| 2D | — | Lr2a, -2b, -2c, -15, -22a, -22b, -39, -41, QTLf | — | Lr54 | — | |
| 3A | — | — | 1562, 9268, 1688 | QTLd | — | — |
| 3B | 0365, 7015, 7142, 5716, 9310, 9170, 6047, 5105, 6802, 5769, 0384, 8096 | Lr27 | — | — | — | — |
| 3D | 1336, 9401 | — | — | Lr24 | — | Lr32 |
| 4A | — | — | 4620 | Lr28 | 5434, 7924 | Lr25 |
| 4B | 1272, 3908 | Lr12, -31 QTLd | Lr30 | — | — | |
| 4D | — | — | — | QTLd | — | — |
| 5A | 0605 | — | — | 4249 | — | |
| 5B | — | Lr52g | 3569, 4996, 5896, 3030, 9598 | Lr18 | 4703 | — |
| 5D | 1400 | — | — | Lr1 | — | — |
| 6A | 7475, 0864, 8006, 7938, 9075 | — | 4229 | — | — | Lr56 |
| 6B | 3130, 4720, 3733 | Lr36,-53 | — | Lr3a, -3bg, -3ka, -9 | — | — |
| 7A | 6034, 8789 | Lr47 | — | Lr20 | 4553 | — |
| 7B | — | — | 7887, 4300, 0600, 7108 | Lr14a, -14b, QTLadf | 9746 | — |
| 7D | 1269, 3328 | Lr29, -34 | — | Lr19 | 0934, 5150, 0366 | — |
The locations of Lr genes as per the USDA–ARS Cereal Disease Lab (http://www.ars.usda.gov/Main/docs.htm?docid=10342) and other publications are shown. The significant DArT markers and Lr genes with unknown location are given in the last two columns. Underlined genes are not present or not known to be present in the lines included in this study.
TABLE 3.
Location of the significant DArT markers associated with stem rust in ESWYT set 1 (ESWYTs 1, 6, 10) and ESWYT set 2 (ESWYTs 20 and 24) using linear mixed model 1 (LMM1) and linear mixed model 2 (LMM2) for each chromosome and the reported Sr genes and QTL
| Short arm (S)
|
Long arm (L)
|
Unknown arm
|
||||
|---|---|---|---|---|---|---|
| Chromosome | DArT (wPt) | Sr gene and QTL | DArT (wPt) | Sr gene and QTL | DArT (wPt) | Sr gene |
| 1A | — | — | — | — | — | — |
| 1B | 8949, 1328, 3465, 4434, 6427, 8986, 1781, 5065, 2614, 5678, 5363, 6777, 5801, 6117, 6833, 8616 | Sr31 | — | Sr14 | 1139, 2019, 1589 | — |
| 1D | — | Sr45 | — | Sr33 | — | Sr18 |
| 2A | — | Sr38 | — | Sr21 | — | Sr32, -34 |
| 2B | 0100, 9402 | Sr19, -23, -36, -40 | — | Sr9a, -9b, -9d, -9e, -9f, -9g, -16, -20, -28 | — | Sr10, -32, -34, -39 |
| 2D | — | Sr6 | — | — | — | — |
| 3A | — | — | 5662 | Sr35 | — | Sr27 |
| 3B | 0365, 0995, 5716, 7015, 8352, 8993, 9170, 5105, 6802, | Sr2, -12 | — | — | — | — |
| 3D | — | — | — | Sr24 | — | — |
| 4A | 2788 | — | 2084, 3795, 7807, 4487 | Sr37 | 5434 | — |
| 4B | 8650 | — | — | Sr7a, -7b | — | — |
| 4D | — | — | — | — | — | Sr41 |
| 5A | — | — | — | — | — | — |
| 5B | — | — | 4996 | — | — | — |
| 5D | — | — | — | Sr30 | — | — |
| 6A | — | Sr8a, -8b | 7623 | Sr13, -26 | — | — |
| 6B | 7662, 1922, 3130, 9990, 6994, 8239, 4720, 3304, 1852, 7777, 3733 | — | 0171, 9256, 1541 | Sr11 | 1541, 5596 | — |
| 7A | — | — | — | Sr15, -22 | 4748 | — |
| 7B | — | — | 1149, 5343, 0600 | Sr17 QTLa | — | — |
| 7D | 2565 | Sr44 | — | Sr25 | 3923, 5674 | |
The locations of Sr genes as per the USDA–ARS Cereal Disease Lab (http://www.ars.usda.gov/Main/docs.htm?docid=10342) and other publications are shown. The significant DArT markers and Sr genes with unknown location are given in the last two columns. Underlined genes are not present or not known to be present in the lines included in this study.
TABLE 4.
Location of the significant DArT markers associated with yellow rust found in ESWYT set 1 (ESWYTs 1, 6, 10) and ESWYT set 2 (ESWYTs 20 and 24) using linear mixed model 1 (LMM1) and linear mixed model 2 (LMM2) in each chromosome and the reported Yr genes and QTL
| Short arm (S)
|
Long arm (L)
|
Unknown arm
|
||||
|---|---|---|---|---|---|---|
| Chromosome | DArT (wPt) | Yr gene and QTL | DArT (wPt) | Yr gene and QTL | DArT (wPt) | Yr gene |
| 1A | 5374, 4765, 2872, 7030, 4029 | — | 3870, 2976, 8016 | — | 6853, 9592 | — |
| 1B | 8949, 3465, 5678, 5801 | Yr9, -10, -24, -26 | 2315 | Yr15, -29 | 1589 | Yr3a, -3b, -3c, -21 |
| 1D | 9380 | — | 7711, 3743 | — | — | Yr25 |
| 2A | 7626, 7901, 6687, 3114 | Yr17, -32 | 2087 | — | 6207 | Yr1 |
| 2B | 0100, 4916, 1489, 9402, 7757 | Yr27, -31 QTLa | 3132, 9350, 7350, 9336, 0047, 3378, 7370, 0049 | Yr5, -7 QTLa | 0950, 4210 | — |
| 2D | — | Yr16 QTLb | 4413 | Yr37 QTLb | 4223 | Yr8 |
| 3A | 9369, 1681 | — | 7756, 7992, 1562, 4725 | — | — | — |
| 3B | 0995, 7142, 9310, 4209, 9510, 5390, 9170, 6047, 5105, 8886, 8238, 8845 | Yr30,Yrns-B1c QTLd | — | — | 2557, 9368 | — |
| 3D | 1336, 9401 | QTLad | — | — | — | — |
| 4A | — | — | 7280, 2084, 3795, 7807, 8271 | — | — | — |
| 4B | 8650, 1046, 6149 | — | 0391 | QTLb | 5265 | — |
| 4D | 4572 | Yr28 | — | QTLb | — | Yr22 |
| 5A | 1165 | QTLa | — | Yr34 | — | — |
| 5B | 5175 | — | 3569, 9467a, 9116 | QTLb | 4418 | Yr19 |
| 5D | — | Yr40 QTLd | — | — | — | — |
| 6A | 7475, 1664, 0832, 4016, 9075 | — | 7623, 9690, 2573, 0959, 7063, 4229, 1642, 7204 | — | — | Yr38 |
| 6B | 7662 | Yr35, -36 QTLb | 1541 | — | 2095 | Yr4a, -4b |
| 7A | 6034, 8789, 4835 | — | 3226, 6495 | — | 4748, 4553 | — |
| 7B | — | Yr6 QTLb | 1149, 3833, 6372, 9925, 5343, 8921, 5547, 5280, 0194, 4300, 6320, 1085, 0504 | Yr39 | 1533 | Yr2 |
| 7D | 1269, 3328 | Yr18 | Yr33 | 0934, 1859, 5049 | — | |
The locations of Yr genes as per the USDA–ARS Cereal Disease Lab (http://www.ars.usda.gov/Main/docs.htm?docid=10342) and other publications are shown. The significant DArT markers and Yr genes with unknown location are given in the last two columns. Underlined genes are not present or not known to be present in the lines included in this study.
TABLE 5.
Location of the significant DArT markers associated with powdery mildew found in ESWYT set 1 (ESWYTs 1, 6, 10) and ESWYT set 2 (ESWYTs 20 and 24) using linear mixed model 1 (LMM1) and linear mixed model 2 (LMM2) for each chromosome and the reported Pm genes and QTL
| Short arm (S)
|
Long arm (L)
|
Unknown arm
|
||||
|---|---|---|---|---|---|---|
| Chromosome | DArT (wPt) | Pm gene and QTL | DArT (wPt) | Pm gene and QTL | DArT (wPt) | Pm gene |
| 1A | 2878 | Pm3a, -3b, -3c, -3d, -3e, -3f, -3g, -3h, -3i, -3j | 2976, 5274, 8016, 0128 | Pm17 QTLab | — | Pm25 |
| 1B | 5678 | Pm8, QTLbc | — | Pm32Pm39d | 3566, 5310 | Pm28 |
| 1D | — | Pm24 | 7711, 3743, 4427, 2897, 1263, 7092 | QTLb | — | Pm10 |
| 2A | — | — | — | Pm4a,-4b | — | — |
| 2B | 0100, 8326 | Pm26 | — | Pm6, Pm33e | 0950 | — |
| 2D | — | — | — | — | 4223 | — |
| 3A | — | — | — | — | — | |
| 3B | 0365, 0995, 8352, 7015, 8983, 4209, 9170, 5105, 8886, 0280, 4412 | QTLf | — | 9368 | — | |
| 3D | 1336, 9401 | Pm13g QTLb | — | — | — | — |
| 4A | — | 8271, 5172, 4487 | QTLf | 5212, 5434, 7821, 7924 | — | |
| 4B | 3908 | Pm7 QTLbhij | — | Pm7 | — | — |
| 4D | — | — | — | — | — | — |
| 5A | 0605 | QTLbc | — | — | 4249 | — |
| 5B | — | Pm30, Pm16m | — | — | — | |
| 5D | — | Pm2 | — | Pm34, -35k | — | — |
| 6A | 7475, 0864, 8006, 7938, 9075 | — | — | Pm21, -31 (MlG) | — | — |
| 6B | 6282, 3116, 3376, 2786 | Pm11, -12, -14, -20 QTLb | 9556 | — | 2095 | Pm27 |
| 7A | 4835 | QTLl | — | Pm1a, -1b, -1c, -1d, -1e, -9Pm37d | 4748 | — |
| 7B | — | — | 4300, 6869 | Pm5a, -5b, -5c, -5d, -5e, Mlxbd | — | — |
| 7D | 1269, 3328 | Pm15,Pm38d | — | Pm29 | 0366, 5049 | Pm19 |
Locations are from Huang and Röder (2004), other publications, and personal communications. The significant DArT markers and Pm genes with unknown location are given in the last two columns.
R. A. McIntosh (personal communication).
TABLE 6.
Location of the significant DArT markers associated with grain yield found in ESWYT set 1 (ESWYTs 1, 6, 10) and ESWYT set 2 (ESWYTs 20 and 24) using linear mixed model 1 (LMM1) and linear mixed model 2 (LMM2) for each chromosome and the reported QTL
| Short arm (S)
|
Long arm (L)
|
Unknown arm
|
||||
|---|---|---|---|---|---|---|
| Chromosome | DArT (wPt) | QTL | DArT (wPt) | QTL | DArT (wPt) | QTL |
| 1A | 5374, 4765, 2872, 9266, 2311, 7030, 4029 | — | 3870, 3904, 5274, 4709, 6754, 8016, 0128 | QTLabc | 9592, 1011, 6853 | QTLd |
| 1B | 8949, 1328, 7094, 3465, 4434, 0974, 6427, 8986, 1781, 5065, 5678, 5363, 6777, 5801, 6117, 6833, 8616 | 1B.1R transl. | 2315, 0944, 2526, 4129 | — | 5316, 1139, 2019 | QTLae |
| 1D | 3738, 9380 | — | 7711, 9857, 3743, 4427, 2897, 1263, 7092 | QTLc | — | QTLbf |
| 2A | 7901, 6687, 9793, 3114 | Ppd3 QTLcg | 5647, 2087 | — | 6207 | QTLefh |
| 2B | 0100, 4916, 6706, 8326, 3983, 5672, 4125, 7757 | Ppd2 | 3132, 9350, 7350, 9336, 2135, 3378, 7360, 0049 | Eps QTLa | 0094, 0950, 4210 | QTLgi |
| 2D | 4144 | Ppd1 Rht8 QTLc | 4413 | Eps | — | QTLefhi |
| 3A | 9369, 1681 | QTLj | 7756, 2698, 7992, 2938, 4725, 9268, 4407, 1688 | — | — | Eps QTLjkl |
| 3B | 0365, 0995, 5716, 7015, 8352, 8983, 7142, 9310, 4209, 9510, 5390, 9170, 6047, 5105, 6802, 8886, 0280, 0384, 8238, 8096, 4412, 8845 | — | — | QTLac | 9368, 2757 | Vrn QTLf |
| 3D | 1365, 9401 | — | — | QTLg | — | Ppd QTLdh |
| 4A | 2788 | QTLcg | 7280, 2084, 3795, 7807, 8271, 4620, 2533 | QTLc | 4660, 7924 | Eps |
| 4B | 3908, 6149 | Rht-1 QTLb | 1505, 8292 | QTLcm | 3608, 5265 | Eps QTLah |
| 4D | QTLg | Rht-2 | — | QTLc | — | Eps QTLd |
| 5A | 1165, 0605 | — | 5231 | Vrn-A1 QTLb | 4249 | QTLabdhno |
| 5B | 6136, 1420, 9666, 5175 | — | 9467, 0103, 4996, 7101, 5896, 3030, 9598 | Vrn-A2 QTLp | 4418, 4703, 4936 | QTLadeq |
| 5D | 1400 | — | — | Vrn-A3 Vrn-4 | — | QTLd |
| 6A | 7475, 0864, 8006, 7938, 9075 | — | 9690, 2573, 0959, 7063, 1642, 7204 | — | — | QTLafr |
| 6B | 9532, 1922, 3130, 9990, 6994, 8239, 4720, 1547, 6282, 3116, 3304, 1852, 7777, 4706, 3733 | — | 6116, 0171, 1264, 3045, 9256, 1541 | — | — | Eps QTLad |
| 7A | 6034, 8789, 4835 | QTLc | 3393, 6495 | QTLbc | 2266, 4553, 4748 | QTLan |
| 7B | — | — | 6273, 9925, 7887, 5343, 8921, 5547, 5280, 0194, 6156, 4300, 7108, 6320, 0504 | QTLb | 1533, 9746 | VrnB4 Vrn5 Eps QTLn |
| 7D | 1269 | — | — | QTLas, 7D.7Ag transl. | 0366, 0934, 3462, 3923, 5049, 5150, 5674, 1859 | QTLf |
Other loci associated with grain yield in wheat are Eps (earliness per se) (Worland 1996; Van Beem et al. 2005), Ppd (photoperiod insensitivity) (Worland 1996; Worland et al. 1998; Van Beem et al. 2005), Rht (semidwarf plant height) (Korzun et al. 1998; Ellis et al. 2002), and Vrn (vernalization) (Worland 1996; Barret et al. 2002; Yan et al. 2003; Van Beem et al. 2005).
DISCUSSION
Association analysis for response to LR:
More than 50 LR resistance genes have been cataloged in bread wheat; some of them have been reported or are known to be present in CIMMYT germplasm, including Lr1, -3, -3bg, -10, -13, -14a, -16, -17a, -19, -21, -23, -24, -26, -34, -42, and -46 and the complementary genes Lr27 and Lr31 (Singh and Mcintosh 1984; Singh and Gupta 1991; Singh and Rajaram 1991; Sayre et al. 1998; Singh et al. 2005) (Table 2). Other known LR genes not known to be present in any of the lines included in this study are listed in Table 2. An objective of the CIMMYT wheat breeding program is to incorporate slow rusting resistance to LR and YR into breeding lines to assure long-term control of these diseases (Rajaram et al. 1988; Singh et al. 2005). Slow rusting resistance is conferred by Lr34, Lr46, and other genes with minor additive effects (Singh et al. 2005). Several studies have identified QTL associated with slow rusting resistance on other chromosomes (such as 7BL) (Table 2).
In this study, many of the DArT markers significantly associated with LR were in the same chromosome regions as previously reported LR resistance genes or previously reported QTL (Table 2). There were also regions accounting for LR resistance variation in which there were no previously known genes or QTL, such as 1AL, 1DL (Figure 2a), 5AS, 5DS (Figure 2e), 6AS, and 6AL (Figure 2f) (Table 2). Chromosome 1B had the largest number of significant markers for LR; some of them may be associated with the reported QTL and genes Lr26 and Lr46. The 1B.1R translocation includes Lr26, YR resistance gene Yr9, SR resistance gene Sr31, and PM resistance gene Pm8; it has also shown an effect on GY (and its components) in several spring wheat cultivars (i.e., Seri M82, Genaro T81, Ures T81, and Veery-related lines) included in this study (Villareal et al. 1991; Singh et al. 1998). Gene Lr46 [pleiotropic or closely linked to Yr29 (William et al. 2003) and associated with PM (Lillemo et al. 2007)] is present in cultivar Pavon 76 (included in this study) and associated with SSR markers gwm259, wmc44, wmc367, and barc80 on 1BL (William et al. 2006). SSR gwm259 is not far from DArT markers wPt0944, wPt2526, and wPt4129 (Figure 2a), which are significant for LR.
Other significant associations include the one between DArT wPt0100 and LR, which appears to relate to Lr16 (Table 2) within the LDb2 of 2BS. This Lr gene has been reported to be linked to wmc661 (McCartney et al. 2005a) and is 1.1 cM from wPt0100 (Figure 2b). Four DArT markers were significantly associated with LR in 7BL (Figure 2g, Table 2) and could be detecting Lr14a present in cultivar Esmeralda M86. In 7DS, the DArT wPt3328 is most likely detecting the slow rusting resistance gene Lr34 that is also linked or pleiotropic to Yr18. Gene Lr34 was present in 60% of CIMMYT bread wheat germplasm in the 1990s (Rajaram et al. 1997) and in several lines of this study (supplemental Table S1 at http://www.genetics.org/supplemental/). Gene Lr19 (linked to Sr25), which is included in a Lophopyrum (Thinopyrum) translocation segment in 7DL was probably not detected due to lack of mapped DArT markers in that arm.
Association analysis for response to SR:
The use of lines with the “Sr2 complex” (Sr2 gene together with other unknown minor genes) in the 1950s provided the foundation for durable resistance to SR (Ortiz et al. 2007a). Of almost 50 known SR resistance genes, Sr2 is the only one conferring slow rusting resistance and is present in Pavon F76 and Veery-related lines (supplemental Table S1 at http://www.genetics.org/supplemental/). Few reports are available on the detection of QTL associated with SR resistance (Table 3).
In this study, several DArT markers were found to be significantly associated with SR resistance (Table 3, Figure 2, a–g). Many map to locations reported from previous linkage mapping studies. In several regions (4BS, 5BL, and 6BS), DArT markers seem to be detecting undesignated SR resistance genes or QTL (Table 3). Several DArT markers in 1BS are most likely associated with the presence of Sr31 of the 1B.1R translocation, which is present in the Veery-related lines included in this study. In 2BS, wPt0100 was found to be associated with SR, GY, LR, and YR in ESWYT set 2, which agrees with the location of Sr23 and is tightly linked to Lr16. Several significant associations for SR were found on chromosome 3BS, where Sr12 and Sr2 are located. Sr2, linked to Lr27, is associated with SSR markers gwm389 and gwm533 (Spielmeyer et al. 2003) and distally located on the short arm where no DArT markers were mapped. It is, however, possible that the DArT markers wPt0365 and wPt0995 are associated with this gene (Table 3, Figure 2c). On chromosome 4AL, three DArT markers in LDb1 and one in LDb2 were associated with SR and could be associated with the presence of Sr7 (Figure 2d).
At the end of 6BL, three DArT markers were associated with SR in ESWYT set 1; they are most likely linked to Sr11, a gene that is in turn linked to Lr3 (McIntosh et al. 1995) that is present in a large number of CIMMYT cultivars (Roelfs and McVey 1979), but to which virulence has evolved in most countries (this is probably why it was not found in ESWYT set 2) (McIntosh et al. 1995). Marker wPt0600 in 7BL could be associated with Sr17, which is found in a wide range of cultivars, including CIMMYT wheat lines, and which is linked to Lr14a and Pm5. Marker wPt3328 in 7DS, corresponding to the position of Lr34/Yr18 (Kerber and Aung 1999), was not significant for SR in this study, despite the fact that Lr34 has previously been associated with enhanced SR resistance.
Association analysis for response to yellow rust (YR):
According to Suenaga et al. (2003), there is significant diversity for genes that have minor to intermediate additive effects on YR resistance. Adult plant resistance to YR and LR in CIMMYT wheat is based on several genes with minor effects, including Yr18 (pleiotropic or closely linked to Lr34), Yr29 (pleiotropic or closely linked to Lr46), and Yr30, which have been deployed to provide a more durable solution against YR (Singh et al. 2005). In addition, of the known seedling YR resistance genes, those reported in CIMMYT wheat lines are, for example, Yr9 (in the 1B.1R translocation), Yr3, Yr1, Yr2, Yr27, Yr7, and Yr6 (McIntosh et al. 1995). In this study, association with YR was found in all chromosomes where DArT markers have been mapped, except at 5D (Table 4). Several significant DArT markers were found in regions where no YR resistance genes or QTL had been reported previously: i.e., 1AS, 1AL, 1DS, 1DL, 2AL, 3AS, 3AL, 4AL, 4BS, 5BS, 6AS, 6AL, 6BL, 7AS, and 7AL (Table 4).
Some significant DArT markers proved to be associated with known YR resistance genes, such as Yr24 at 1B, which is allelic to Yr26 and closely linked to barc187 (Li et al. 2006) (Figure 2a). Boukhatem et al. (2002) identified two major QTL for durable YR resistance on chromosome 2B, of which one was present in Opata 85 and mapped near the SSR marker gwm410. This marker is also present in LDb3 with wPt7757, which is significantly associated with YR (Figure 2b, Table 4). On 2DL, a QTL associated with a reduction in YR infection type was reported to be linked to marker gwm349 (Mallard et al. 2005), which in our study is ∼3 cM from wPt4413 (Figure 2b, Table 4). In addition, Suenaga et al. (2003) detected a QTL associated with YR resistance close to marker gwm349 on 2DL (Figure 2b) within LDb6 near DArT marker wPt4413. Yr30, at 3BS, has been reported to be linked to Sr2 and Lr27, and to SSR marker gwm533 (Spielmeyer et al. 2003), and may be marked by wPt0995 and other DArT markers at LDb3 (Figure 2c) (it is present in Opata 85 and Pavon F76, included in this study). Singh et al. (2000) and Boukhatem et al. (2002) identified a region on chromosome 3DS associated with YR in Opata 85, which could have been identified by wPt1336 and wP19401.
A QTL for adult plant YR resistance in 4BL was linked to SSRs wmc48c and gwm513 (Suenaga et al. 2003). In our study, marker wPt6149 was significantly associated with YR and located between these two markers (Figure 2d). A QTL for adult plant YR resistance in 4D was associated with marker psp3103 (Suenaga et al. 2003), which is 8.4 cM from wPt4572 that is significant for YR (Figure 2d). The same authors identified a QTL for YR resistance in 5BL between SSRs gwm335 and gwm777, where several DArT markers were significantly associated with YR (Table 4, Figure 2e) (see supplemental Figure S15 at http://www.genetics.org/supplemental/ for the location of gwm777 on the map including all markers). The only reported gene on chromosome 6A is Yr38, which was recently transferred into wheat from Aegilops sharonensis (Marais et al. 2006). In contrast, this study found several linked and unlinked DArT markers significantly associated with YR on chromosome 6A in both ESWYT sets. These results indicate the presence of unknown YR resistance genes or alleles on this chromosome. On 7B, Suenaga et al. (2003) identified a QTL linked to SSRs gwm333 and gwm46, which in our study are located at LBb4 near wPt1149, wPt3833, and wPt6372, all of which were significantly associated, among others, to YR (Table 4, Figure 2g). At 7D, the adult plant resistance gene Yr18 is most likely being detected by DArT wPt3328 or wPt1100 (Figure 2g).
Association analysis for response to PM:
CIMMYT bread wheat germplasm is generally susceptible to PM due to the lack of natural epidemics of B. graminis f.sp. tritici in Mexico. Hence, there has not been direct selection for resistance against this pathogen in the breeding program. There is, however, considerable genetic variation for PM resistance in CIMMYT germplasm, as indicated by the many significant DArT markers associated with the trait in this study. Most of the significant chromosomal areas for PM resistance were associated with known Pm genes or QTL (Table 5). For example, the significant markers on chromosome arms 1BS, 1DL, 2BS, 3DS, 6AS, and 7BL correspond to the locations of Pm8, Pm24, Pm26, Pm13, Pm31, and Pm5, respectively, and the groups of significant markers for PM resistance on 1AL, 3BS, 4AL, 6BS, and 7DS correspond to reported QTL (Table 5). Recently, Nematollahi et al. (2007) detected PM resistance allele Pm5d at 7BL near SSRs gwm577 and wmc 581. These two SSRs are located in the same LDb6 of 7BL on our map (Figure 2g), where wPt6869, significant for PM, was found (and near wPt4300, significant for PM). There were also many significant DArT markers located in areas where no Pm gene or QTL had been reported previously, e.g., wPt2872 on 1AS, wPt4427-wPt7092 on 1DL, wPt4412 on 3BL, wPt9256 on 6BL, and wPt1085-wPt6869 on 7BL.
It is interesting to note that all the chromosomal areas that showed a significant effect on PM in this study also harbored genetic factors for resistance to at least one of the three rust diseases. This indicates that the PM resistance at these loci might have been indirectly selected for by selection for LR or YR. For example, the race-specific resistance gene Pm8 is co-inherited with Sr31, Lr29, and Yr9 in the 1B.1R translocation, and the Pm5 locus on 7BL is linked to Sr17 and Lr14. Such co-inheritance of resistance to PM and the rust diseases is also evident for race nonspecific adult plant resistance. For example, the Lr34/Yr18 locus on 7DS has been shown to confer partial resistance to PM (Spielmeyer et al. 2005; Lillemo et al. 2007), as does the Lr46/Yr29 locus on 1BL (Lillemo et al. 2007). These genes have been designated Pm38 and Pm39, respectively (R. A. McIntosh, personal communication). The DArT marker wPt3328 appears to be associated with Pm38, whereas this analysis failed to detect any effect of Pm39 on 1BL.
Association analysis for GY:
The genetics of yield and yield components in bread wheat are complex in nature and controlled by a large number of major and minor QTL (Kumar et al. 2007). Dispersion of genes affecting GY across the wheat genome appears to be a characteristic feature (Laperche et al. 2007) that was also observed in this study (Table 6). In this study, 213 DArT markers (or 88% of the mapped markers) were significantly associated with GY. About 58% of the significant DArT markers for GY mapped to the B genome, 31% to the A genome, and 11% to the D genome. A large proportion of significant DArT markers for GY were located on chromosomes 1B, 3B, and 6B, and no DArT markers on chromosome 4D were significantly associated with GY. Many of the DArT markers found to be significantly associated with GY map to regions previously reported for this trait (see discussion below). This study suggests that chromosomes 1AS, 1BS, 1BL, 1DS, 2AL, 3AL, 3BS, 5AS, 5DS, 6AS, 6AL, 6BS, 6BL, and 7DS (Table 6, Figure 2, a–g) contain regions affecting GY that have not been reported previously. Some of these regions may provide increases in GY due not only to additive effects but also to additive × additive interaction effects.
The results of this study, highlighting chromosomal regions affecting GY, are supported by results of previous studies, including the following:
One major allele for increased GY was identified on chromosome 2B near gwm410. This yield effect may be linked to a QTL for stay green found on 2B, which has been shown to improve yield under both drought stress and optimal conditions (Verma et al. 2004). In the present study, in ESWYT set 2, GY was significantly associated with markers wpt5672, wPt4125, and wPt7757 in LDb3 (where gwm410 is located) (Figure 2b).
Kumar et al. (2007) found consistent QTL on 2DS linked to SSR gwm261 for five yield traits. In our study, marker gwm261 was found in LDb6 of 2D at ∼2.20 cM from wPt4144, which was associated with yield in ESWYT set 1 (Figure 2b).
Dilbirligi et al. (2006) found that marker barc12 on chromosome arm 3AS was associated with GY. In our study, barc12 was positioned at 3AL but located near wPt4725 and wPt9262, which were significant for GY, YR, and LR. Also in the present study, five DArT markers in LDb1 on chromosome arm 3AS (Figure 2c) were associated with GY.
On chromosome 7, Kumar et al. (2007) identified two regions associated with GY at 7AS (interval gwm130–gwm1171) and at 7AL (interval gwm332–gwm698). The region at 7AS and marker gwm130 coincides with the location of wPt6034 and wPt8789 (LDb1), which were significant for GY, and the region in 7AL within LDb2 contained gwm698 and also wPt6495, which was significant for GY in ESWYT set 1 (Figure 2g).
A QTL for GY was mapped to 4AS near marker wmc48 (McCartney et al. 2005b). Our complete map (supplemental Figure S4 at http://www.genetics.org/supplemental/) shows wmc48 to be located 13.4 cM from wPt2788 (Figure 2d) and significantly associated with GY.
Clusters of QTL for GY and yield components mapped to 1D, 2A, 6B, and 7D in Li et al. (2007). DArT marker wPt3738 on 1D in our map is significantly associated with GY; however, this region in our map is closer to SSR gwm642 than was reported by Li et al. (2007). Li's cluster of QTL at 2A may coincide with wPt7901, wPt6687, and wPt9793 at 2AS (Figure 2c), and Li's QTL at 6B is near gwm132, which in Figure 2c is ∼20 cM from several significant DArT markers for GY. Finally, the QTL at 7D seems to be distally associated with wPt1269 at 7DS in our map; nevertheless, this may be due to the relatively small number of DArT markers mapping to this chromosome.
Lines carrying the 1B.1R translocation show increased GY (Carver and Rayburn 1994), which may explain the significant associations found between GY and DArT markers mapping to the long arm of chromosome 1B (Figure 2a). The 7DL.7Ag translocation segment, associated with GY increase under irrigated conditions and GY decrease in unfavorable environments (Braun et al. 1996; Singh et al. 1998), was not detected in this study. CIMMYT wheat lines are selected for consistently high performance under many diverse growing conditions and so may not have positive selection either for or against this translocation.
The short-stature wheats containing dwarfing Rht genes have been associated with high GY (Trethowan et al. 2007). Our study shows significant associations with GY for known chromosomal regions bearing the dwarfing alleles Rht-B1 and Rht8 genes (Ortiz et al. 2007b). (Table 6). While all CIMMYT semidwarf bread wheats are expected to carry Rht-B1b or Rht-D1b, it was shown that a height-promoting allele at the Rht8 locus is also frequently present in CIMMYT wheats, and it is speculated that this allele might confer an adaptive advantage by partially counteracting the dramatic effects of Rht-B1b and Rht-D1b (Worland et al. 2001).
The importance for GY of earliness per se (Eps), photoperiod insensitivity (Ppd), and vernalization (Vrn) genes was indicated by our study. The anthesis rate in wheat also depends on the variation in Eps, Ppd, and Vrn genes (Van Beem et al. 2005). Vrn genes are common in CIMMYT bread wheat germplasm, whereas Eps genes occur only at low frequencies. Chromosomal regions bearing these genes were significantly associated with QTL for GY, as revealed by DArT markers in both ESWYT sets included in this study (Table 6).
Association between all traits:
Markers for many of the traits studied here were found to be colocalized in the same genomic regions, as well as often significantly associated. Multiple regression of GY on the disease traits shows that only variability for LR and PM significantly contributes to GY variability, with a coefficient of determination of 22%. Significant (P ≤ 0.05) rank correlations (rS) were found for the following pairs of traits across the ESWYTs: GY and YR (rS = −0.23), SR and YR (rS = 0.45), LR and YR (rS = 0.40), LR and PM (rS = 0.37), and YR and PM (rS = 0.34). These results indicate that the YR-resistant wheat lines included in this study were often resistant to SR, LR, and PM as well. In addition, the lines that showed the highest susceptibility to YR also displayed lower GY due to a significant negative correlation. In ESWYTs 1 and 6, significant associations were found between SR and YR (rS = 0.60 and 0.55, respectively). In ESWYT 6, associations were also found between SR and PM (rS = 0.40) and YR and PM (rS = 0.46). In ESWYT 10, a significant rank correlation was found between SR and YR (rS = 0.46). In ESWYT 20, significant associations were found between GY and YR (rS = −0.40), GY and PM (rS = −0.43), SR and LR (rS = 0.34), and LR and YR (rS = −0.47). In ESWYT 24, significant associations were found between LR and PM (r = 0.35) and LR and YR (rS = 0.35) (data not shown).
Several DArT markers were associated with more than one trait investigated in our study. Several resistance genes (R genes) affecting different diseases have been reported to be clustered in specific regions of the genome, such as Lr20-Sr15-Pm1 (Neu and Keller 2002). These clusters of R genes are important breeding targets, because the transfer of such a region results in resistance to several pathogens simultaneously (Neu and Keller 2002). Pleiotropy or linkage between adult plant resistance genes for different pathogens has also been reported. For example, Lr34 is completely linked or pleiotropic to Yr18, which is also linked to a gene conditioning tolerance to barley yellow dwarf virus (Bydv1) (Singh 1993) and to PM resistance (Spielmeyer et al. 2005; Lillemo et al. 2007). In this study, associations between disease resistance and GY were probably caused by the reduction in yield loss due to susceptibility, since only variability for LR and PM significantly contributed to GY variability, explaining 22% of GY variation. This result indicates that ESWYT materials maintained a relatively high level of overall disease resistance, with abiotic factors presumably accounting for most of the variation.
Implicit marker effects in statistical models:
The fixed effects of markers are implicitly considered when comparing the BLUPs of the lines obtained from LMM1 by using the variance–covariance matrix of the BLUPs. Modeling the marker effects in this manner has two advantages over the model that explicitly includes markers as fixed effects: (1) information contained in relationship matrix A is fully exploited, as its effect is reflected in the BLUPs of the lines (and in their variance–covariance matrix) and, thus, in the contrast for each marker in each subpopulation and across subpopulations; and (2) LMM1 can be fitted to avoid nonidentification and overparameterization. This last issue is more of a problem when some lines lack data for some markers. Another point worth mentioning is that since markers represent attributes of lines, an appropriate model would include lines nested within markers (Malosetti et al. 2007) and within subpopulations. When markers are explicitly included in LMM1 and LMM2 as fixed (or random) effects, it is not clear how the effect of relationship matrix A (or kinship matrix K, in case markers are being used) would be reflected in the contrasts of the BLUPs of the lines in each subpopulation. Furthermore, aliasing problems due to ambiguities in the model are often encountered when including (1) markers as fixed (or random) effects, (2) the use of relationship matrix A for modeling GE, and (3) information from the Q matrix (as fixed or random covariables). Marker effects and Q matrix values are related; therefore, it would be advisable to calculate submatrices A for each marker and each subpopulation in matrix Q.
A statistical model that explicitly includes subpopulations, markers, and lines must consider (1) the main effect of markers and the interaction of marker × environment, (2) the main effect of subpopulations and its interaction with environments, and (3) the main effects of lines nested within subpopulations and markers and their interaction with environments. Interaction effects can be estimated separately or combined with principal effects (i.e., markers could be estimated separately or combined with marker × environment interaction). Nevertheless, this implies that for each marker type only the COP (or kinship K) of the lines included at each level of the marker must be considered, and lines with no markers must not be included in the model. Without clear separation and definition of these terms, ambiguity in the linear mixed model will lead to aliasing in the estimation process. Model LMM1 draws on an independent source of information for measuring the covariance among relatives (from matrix A) and uses information on population structure (from matrix Q) only as weights in the contrast of the BLUPs of the lines for each marker. When information from Q is included in the model as fixed covariates, then marker effects are estimated (or predicted) at a certain value of the covariates; however, it is not clear what that value is.
The models used in this study include the relationship matrix and the population structure of each ESWYT and of the two ESWYT sets. Yu et al. (2006) found that incorporating the outcome of population structure contained in the Q matrix increases the power to detect true marker–trait association. In addition, Piepho (2005) shows that genetic correlations (for QTL and QTL × environment interaction) in multienvironment trials lead to a substantial increase of the type I error rate when testing for QTL effects. On the basis of these findings, we can conclude that, by using LMM1 and LMM2, the risk of finding false marker–trait associations was reduced. The application of appropriate statistical models for assessing GE is useful in plant breeding (Sorrells 2007) and should be adopted in association analysis studies. Statistical models other than LMM1 and LMM2 should be further studied.
Conclusions:
Results from this study show DArT markers significantly associated with the measured traits in chromosomal regions where genes or QTL have been previously reported, as well as significantly associated DArT markers in regions where neither genes nor QTL have previously been reported for these traits. The role of these regions will need to be further investigated. The consensus map used in this study has proven to be sufficiently precise to compare the locations of DArT markers from this study with those of other markers (e.g., SSRs) previously reported. Several of the known cataloged genes, such as Lr47, were recently transferred from alien or related species and thus are not expected to be present in the material included in this study. However, it cannot be excluded that noncataloged corresponding homologs of these alien loci could be present in the wheat lines investigated in this study, and that they could have some effect in reducing disease severity. For example, the markers on 7AS could be tagging a homolog of the Lr47 locus rather than a completely new LR resistance gene. It is also likely that many of these gene-rich regions will carry previously unidentified minor slow rusting or adult plant resistance genes.
The historical phenotypic data used in this study were collected from many different environments worldwide, which led to GE effects being observed for all traits. Partitioning the total genetic effect into additive and additive × additive effects used in association analysis could be an added advantage for identifying significant markers associated with phenotypic traits with these effects, which will facilitate further genetic enhancement of the crop.
Concerning the disease traits, the variation of the pathogen races occurring at different locations is likely to reduce the identification of race-specific resistance. Most of the known cataloged genes are race specific and effective only in some geographic areas. In this study, these pathogens may not have been present at high frequencies in the years of evaluation at many locations. For example, Lr3a, which occurs in several CIMMYT wheat lines, could not be identified in this study (at least in ESWYT set 2) because virulence to this gene is common worldwide. Virulence to Lr1 is also common in most wheat-growing areas, and the gene would have been difficult to detect even if more markers had mapped to the chromosome containing Lr1. Loss of effectiveness due to the presence of virulent races is probably the reason why we could not detect the chromosomal regions of some genes known to be present in CIMMYT wheat materials. Only a few genes, such as Lr34/Yr18, Lr46/Yr29, and Yr30/Sr2, are nonrace specific in nature and should have small-to-intermediate effects across different environments. Analyses did, in fact, identify chromosomal regions carrying the above genes. Finally, differences and similarities between ESWYT sets 1 and 2 track genetic changes in the CIMMYT wheat breeding program over the past 24 years, and further analyses will shed greater light on the genetic determinants of GY and disease resistance in an evolving context.
Acknowledgments
The authors thank the numerous cooperators in national agricultural research institutes who carried out the elite spring wheat yield trials and provided the phenotypic data analyzed in this article plus the International Maize and Wheat Improvement Center's international nursery and seed distribution units who prepared and distributed the seed and computerized the data. We thank Alma McNab for the English editing.
References
- Akbari, M., P. Wenzl, V. Caig, J. Carling, L. Xia et al., 2006. Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theor. Appl. Genet. 113: 1409–1420. [DOI] [PubMed] [Google Scholar]
- Ali, M. L., P. S. Baenziger, A. A. Zakaria, M. Shafqat, K. S. Gill et al., 2007. Mapping QTLs for GY and other agronomic traits located on chromosome 3A of wheat. Abstracts of Plant and Animal Genome XV Conference, January 13–17, 2007, San Diego. (http://www.intl-pag.org/15/abstracts/PAG15_P05c_273.html).
- Arbelbide, M., and R. Bernardo, 2006. Mixed-model QTL mapping for kernel hardness and dough strength in bread wheat. Theor. Appl. Genet. 112: 885–890. [DOI] [PubMed] [Google Scholar]
- Barret, B., M. Bayram and K. Kidwell, 2002. Identifying AFLP and microsatellite markers for vernalization response gene Vrn-B1 in hexaploid wheat using reciprocal mapping populations. Plant Breed. 121: 400–406. [Google Scholar]
- Benjamini, Y., and Y. Hochberg, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57(1): 289–300. [Google Scholar]
- Börner, A., E. Schumann, A. Fürste, H. Cöster, B. Leithold et al., 2002. Mapping of quantitative trait loci determining agronomic important characters in hexaploid wheat (Triticum aestivum L.). Theor. Appl. Genet. 105: 921–936. [DOI] [PubMed] [Google Scholar]
- Bougot, Y., J. Lemoine, M. T. Pavoine, H. Guyomar'ch, V. Gautier et al., 2006. A major QTL effect controlling resistance to powdery mildew in winter wheat at the adult plant stage. Plant Breed. 125: 550–556. [Google Scholar]
- Boukhatem, N., P. V. Baret, D. Mingeot and J. M. Jacquemin, 2002. Quantitative loci for resistance against yellow rust in two wheat-derived recombinant inbred line populations. Theor. Appl. Genet. 104: 111–118. [DOI] [PubMed] [Google Scholar]
- Braun, H.-J., S. Rajaram and M. Van Ginkel, 1996. CIMMYT's approach to breeding for wide adaptation. Euphytica 92: 175–183. [Google Scholar]
- Breseghello, F., and M. E. Sorrells, 2006. a Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 172: 1165–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breseghello, F., and M. E. Sorrells, 2006. b Association analysis as a strategy for improvement of quantitative traits in plants. Crop Sci. 46: 1323–1330. [Google Scholar]
- Burgueño, J., J. Crossa, P. L. Cornelius, R. Trethowan, G. McLaren et al., 2007. Modeling additive × environment and additive × additive × environment using genetic covariances of relatives of wheat genotypes. Crop Sci. 47: 311–320. [Google Scholar]
- Campbell, B. T., P. S. Baenziger, K. S. Gill, K. M. Eskridge, H. Budak et al., 2003. Identification of QTLs and environmental interactions associated with agronomic traits on chromosome 3A of wheat. Crop Sci. 43: 1493–1505. [Google Scholar]
- Campbell, B. T., P. S. Baenziger, K. M. Eskridge, H. Budak, N. A. Streck et al., 2004. Using environmental covariates to explain genotype × environment and QTL × environment interactions for agronomic traits on chromosome 3A of wheat. Crop Sci. 44: 620–627. [Google Scholar]
- Carver, B. F., and A. L. Rayburn, 1994. Comparison of related wheat stocks possessing 1B or 1RS.1BL chromosomes: agronomic performance. Crop Sci. 34: 1505–1510. [Google Scholar]
- Cenci, A., R. D'Ovidio, O. A. Tanzarella, C. Ceoloni and E. Porceddu, 1999. Identification of molecular markers linked to Pm13, an Aegilops longissima gene conferring resistance to powdery mildew in wheat. Theor. Appl. Genet. 98: 448–454. [Google Scholar]
- Chantret, N., D. Mingeot, P. Sourdille, M. Bernard, J. M. Jacquemin et al., 2001. A major QTL for powdery mildew resistance is stable over time and at two development stages in winter wheat. Theor. Appl. Genet. 103: 962–971. [Google Scholar]
- Chen, X. M., Y. H. Luo, X. C. Xia, L.Q. Xia, X. Chen et al., 2005. Chromosomal location of powdery mildew resistance gene Pm16 in wheat using SSR marker analysis. Plant Breed. 124: 225–228. [Google Scholar]
- Cockerham, C. C., 1971. Higher order probability functions of identity of alleles by descent. Genetics 69: 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossa, J., J. Burgueño, P. L. Cornelius, G. McLaren, R. Trethowan et al., 2006. Covariances of relatives for predicting breeding values of wheat genotypes. Crop Sci. 46: 1722–1733. [Google Scholar]
- Cullis, B. R., F. M. Thomson, J. A. Fisher, A. R. Gilmour and R. Thompson, 1996. The analysis of the NSW wheat variety database. II. Variance component estimation. Theor. Appl. Genet. 92: 28–39. [DOI] [PubMed] [Google Scholar]
- Dilbirligi, M., M. Erayman, B. T. Campbell, H. S. Randhawa, P. S. Baenziger et al., 2006. High density mapping and comparative analysis of agronomically important traits on wheat chromosome 3A. Genomics 88: 74–87. [DOI] [PubMed] [Google Scholar]
- Ellis, M. H., W. Spielmeyer, K. R. Gale, G. J. Rebetzke and R. A. Richards, 2002. “Perfect”markers for the Rht-B1b and Rht-D1b dwarfing genes in wheat. Theor. Appl. Genet. 105: 1038–1042. [DOI] [PubMed] [Google Scholar]
- Faris, J. D., W. L. Li, D. J. Liu, P. D. Chen and B. S. Gill, 1999. Candidate gene analysis of quantitative disease resistance in wheat. Theor. Appl. Genet. 98: 219–225. [Google Scholar]
- Flint-Garcia, S. A., J. M. Thornsberry and E. S. Blucker, IV, 2003. Structure of linkage disequilibrium in plants. Annu. Rev. Plant. Biol. 54: 357–374. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Hernandez, J. L., E. M. Elias and S. F. Kianian, 2004. Mapping genes for grain protein concentration and GY on chromosome 5B of Triticum turgidum (L.) var. dicoccoides. Euphytica 139: 217–225. [Google Scholar]
- Groos, C., N. Robert, E. Bervas and G. Charmet, 2003. Genetic analysis of grain-protein content, GY, and thousand-kernel weight in bread wheat. Theor. Appl. Genet. 106: 1032–1040. [DOI] [PubMed] [Google Scholar]
- Hiebert, C., J. Thomas and B. McCallum, 2005. Locating the broad-spectrum wheat leaf rust resistance gene Lr52 (LrW) to chromosome 5B by a new cytogenetic method. Theor. Appl. Genet. 110: 1453–1457. [DOI] [PubMed] [Google Scholar]
- Huang, X. Q., and M. S. Röder, 2004. Molecular mapping of powdery mildew resistance genes in wheat: a review. Euphytica 137: 202–223. [Google Scholar]
- Huang, X. Q., H. Cöster, M. W. Ganal and M. S. Röder, 2003. Advanced backcross QTL analysis for the identification of quantitative trait loci alleles from wild relatives of wheat (Triticum aestivum L.). Theor. Appl. Genet. 106: 1379–1389. [DOI] [PubMed] [Google Scholar]
- Huang, X. Q., H. Kempf, M. W. Ganal and M. S. Röder, 2004. Advanced backcross QTL analysis in progenies derived from a cross between a German elite winter wheat variety and a synthetic wheat (Triticum aestivum L.). Theor. Appl. Genet. 109: 933–943. [DOI] [PubMed] [Google Scholar]
- Huang, X. Q., S. Cloutier, L. Lycar, N. Radovanovic, D. G. Humphreys et al., 2006. Molecular detection of QTLs for agronomic and quality traits in a double haploid population derived from two Canadian wheats (Triticum aestivum L.). Theor. Appl. Genet. 113: 753–766. [DOI] [PubMed] [Google Scholar]
- Jakobson, I., H. Peusha, L. Timofejeva and K. Jarve, 2006. Adult plant and seedling resistance to powdery mildew in a Triticum aestivum x Triticum militinae hybrid line. Theor. Appl. Genet. 112: 760–769. [DOI] [PubMed] [Google Scholar]
- Jannink, J. L., and J. B. Walsh, 2002. Association mapping in plant populations, pp. 59–68 in Quantitative Genetics Genomics and Plant Breeding, edited by M. S. Kang. CABI, Wallingford, UK.
- Kato, K., H. Miura and S. Sawada, 2000. Mapping QTLs controlling grain yield and its components on chromosome 5A of wheat. Theor. Appl. Genet. 101: 1114–1121. [Google Scholar]
- Keller, M., B. Keller, G. Schachermayr, M. Winzeler, J. E. Schmid et al., 1999. Quantitative trait loci for resistance against powdery mildew in a segregating wheat x spelt population. Theor. Appl. Genet. 98: 903–912. [Google Scholar]
- Kempthorne, O., 1969. An Introduction to Genetics Statistics. Iowa State University Press, Ames, IA.
- Kerber, E. R., and T. Aung, 1999. Leaf rust resistance gene Lr34 associated with nonsuppression of stem rust resistance in the wheat cultivar Canthatch. Phytopathology 89: 518–521. [DOI] [PubMed] [Google Scholar]
- Kirigwi, F. M., M. Van Ginkel, G. Brown-Gedira, B. S. Gill, G. M. Paulsen et al., 2007. Markers associated with a QTL for grain yield in wheat under drought. Mol. Breed. 20: 401–413. [Google Scholar]
- Korzun, V., M. S. Röder, M. W. Ganal, A. J. Worland and C. N. Law, 1998. Genetic analysis of the dwarfing gene (Rht8) in wheat. Part I. Molecular mapping of Rht8 on the short arm of chromosome 2D of bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 98: 1104–1109. [Google Scholar]
- Kraakman, A. T. W., R. E. Niks, P. M. M. M. Van den Berg, P. Stam and F. A. van Eeuwijk, 2004. Linkage disequilibrium mapping of the yield stability in modern spring barley cultivars. Genetics 168: 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, N., P. L. Kulwal, H. S. Balyan and P. K. Gupta, 2007. QTL mapping for yield and yield contribution traits in two mapping populations of bread wheat. Mol. Breed. 19: 163–177. [Google Scholar]
- Laperche, A., M. Brancourt-Hulme, E. Heumez, O. Gardet, E. Hanson et al., 2007. Using genotype x nitrogen interaction variables to evaluate the QTL involved in wheat tolerance to nitrogen constraints. Theor. Appl. Genet. 115: 399–415. [DOI] [PubMed] [Google Scholar]
- Li, G. Q., Z. F. Li, W. Y. Yang, Y. Zhang, Z. H. He et al., 2006. Molecular mapping of stripe rust resistance gene YrCH42 in Chinese wheat cultivar Chuanmai 42 and its allelism with Yr24 and Yr26. Theor. Appl. Genet. 112: 1434–1440. [DOI] [PubMed] [Google Scholar]
- Li, S., J. Jia, X. Wei, X. Zhang, L. Li et al., 2007. A intervarietal genetic map and QTL anaysis for yield traits in wheat. Mol. Breed. 20: 167–168. [Google Scholar]
- Liang, S. S., K. Suenaga, Z. H. He, Z. L. Wang, H.Y. Liu et al., 2006. Quantitative trait loci mapping for adult-plant resistance to powdery mildew in bread wheat. Phytopathology 96: 784–789. [DOI] [PubMed] [Google Scholar]
- Lillemo, M., Å. Bjørnstad, B. Asalf, R. P. Singh, J. Huerta-Espino et al., 2006. Identification of molecular markers associated with partial resistance to powdery mildew, leaf rust and stripe rust in bread wheat line Saar. EUCARPIA Cereals Section Meeting, November, 2006, Lleida, Spain.
- Lillemo, M., R. P. Singh, J. Huerta-Espino, X. M. Chen, Z. H. He et al., 2007. Leaf rust resistance gene Lr34 is involved in powdery mildew resistance of CIMMYT bread wheat line Saar, pp. 97–102 in Wheat Production in Stressed Environments (Developments in Plant Breeding, Vol. 12), edited by H. T. Buck, J. E. Nisi and N. Salomon. Proceedings of the 7th International Wheat Conference, November 27–December 2, 2005, Mar del Plata, Argentina.
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- Mallard, S., D. Gaudet, A. Aldeia, C. Abelard, A. L. Besnard et al., 2005. Genetic analysis of durable resistance to yellow rust in bread wheat. Theor. Appl. Genet. 110: 1401–1409. [DOI] [PubMed] [Google Scholar]
- Malosetti, M., C. G. Van der Linden, B. Vosman and F. E. Van Eeuwijk, 2007. A mixed-model approach to association mapping using pedigree information with an illustration of resistance to Phytophthora infestans in potato. Genetics 175: 879–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais, G. F., B. McCallum and A. S. Marais, 2006. Leaf rust and stripe rust resistance genes derived from Aegilops sharonensis. Euphytica 149: 373–380. [Google Scholar]
- Marza, F., G.-H. Bai, B. F. Carver and W.-C. Zhou, 2006. Quantitative trait loci for yield and related traits in the wheat population. Ning7840 × Clark. Theor. Appl. Genet. 112: 688–698. [DOI] [PubMed] [Google Scholar]
- McCartney, C. A., D. J. Somers, B. D. McMcallum, J. Thomas, D. J. Humphrey et al., 2005. a Microsatellite tagging of the leaf rust resistance gene Lr16 on wheat chromosome 2BSc. Mol. Breed. 15: 329–337. [Google Scholar]
- McCartney, C. A., D. J. Somers, D. G. Humphreys, O. Lukow, N. Ames et al., 2005. b Mapping quantitative trait loci controlling agronomic traits in the spring wheat cross RL4452 × ‘AC Domain’. Genome 48: 870–883. [DOI] [PubMed] [Google Scholar]
- McIntosh, R. A., C. R. Wellings and R. F. Park, 1995. Wheat Rusts: An Atlas of Resistance Genes. CSIRO, Melbourne, Australia
- McLaren, C. G., R. M. Bruskiewich, A. M. Portugal and A. B. Cosico, 2005. The International Rice Information System. A platform for meta-analysis of rice crop data. Plant Physiol. 139: 637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messmer, M. M., R. Seyfarth, M. Keller, G. Schachermayr, M. Winzeler et al., 2000. Genetic analysis of durable leaf rust resistance in winter wheat. Theor. Appl. Genet. 100: 419–431. [Google Scholar]
- Mingeot, D., N. Chantret, P. V. Baret, A. Dekeyser, N. Boukhatem et al., 2002. Mapping QTL involved in adult plant resistance to powdery mildew in the winter wheat line RE714 in two susceptible genetic backgrounds. Plant Breed. 121: 133–140. [Google Scholar]
- Miranda, L. M., J. P. Murphy, D. Marshall and S. Leath, 2006. Pm34: a new powdery mildew resistance gene transferred from Aegilops tauschii Coss. to common wheat (Triticum aestivum L.). Theor. Appl. Genet. 113: 1497–1504. [DOI] [PubMed] [Google Scholar]
- Miranda, L. M., J. P. Murphy, D. Marshall, C. Cowger and S. Leath, 2007. Chromosomal location of Pm35, a novel Aegilops tauschii derived powdery mildew resistance gene introgressed into common wheat (Triticum aestivum L.). Theor. Appl. Genet. 114: 1451–1456. [DOI] [PubMed] [Google Scholar]
- Miura, H., B. B. Parker and J. W. Snape, 1992. The location of major genes and associated quantitative trait loci on chromosome arm 5BL of wheat. Theor. Appl. Genet. 85: 197–204. [DOI] [PubMed] [Google Scholar]
- Nelson, J. C., R. P. Singh, J. E. Autrique and M. E. Sorrells, 1997. Mapping genes conferring and suppressing leaf rust resistance in wheat. Crop Sci. 37: 1928–1935. [Google Scholar]
- Nematollahi, G., V. Mohler, G. Wenzel, F. J. Zeller and S. L. K. Hsam, 2007. Microsatellite mapping of powdery mildew resistance allele Pm5d from common wheat line IGVI-455. Euphytica (in press).
- Neu, C. N. S., and B. Keller, 2002. Genetic mapping of the Lr20-Pm1 resistance locus reveals suppressed recombination on chromosome arm 7AL in hexaploid wheat. Genome 45: 737–744. [DOI] [PubMed] [Google Scholar]
- Oakey, H., A. A. Verbyla, W. Pitchford, B. Cullis and H. Kuchel, 2006. Joint modeling of additive and non-additive genetic line effects in single field trials. Theor. Appl. Genet. 113(5): 809–819. [DOI] [PubMed] [Google Scholar]
- Ortiz, R., D. Mowbray, C. Dowswell and S. Rajaram, 2007. a Norman E. Borlaug: the humanitarian plant scientist who changed the world. Plant Breed. Rev. 28: 1–37. [Google Scholar]
- Ortiz, R., R. Trethowan, G. Ortiz-ferrara, M. Iwanaga, J. H. Dodds et al., 2007. b High yield potential, shuttle breeding, genetic diversity, and a new international wheat improvement strategy. Euphytica 157: 365–384. [Google Scholar]
- Parisseaux, B., and R. Bernardo, 2004. In silico mapping of quantitative trait loci in maize. Theor. Appl. Genet. 109: 508–514. [DOI] [PubMed] [Google Scholar]
- Perrier, X., A. Flori and F. Bonnot, 2003. Data analysis methods, pp. 43–76 in Genetic Diversity of Cultivated Tropical Plants, edited by P. Hamon, M. Seguin, X. Perrier and J.-C. Glaszmann. Enfield Science Publishers, Plymouth, UK.
- Piepho, H.-P., 2005. Statistical tests for QTL and QTL-by-environment effects in segregating populations derived from line crossa. Theor. Appl. Genet. 110: 561–566. [DOI] [PubMed] [Google Scholar]
- Pritchard, J. K., M. Stephen and P. Donnely, 2000. Inference on population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarrie, S. A., A. Steed, C. Calestani, A. Semikhodskii, C. Lebreton et al., 2005. A high density genetic map of hexaploid wheat (Triticum aestivum L.) from the cross Chinese Spring × SQ1 and its use to compare QTLs for grain yield across a range of environments. Theor. Appl. Genet. 110: 865–880. [DOI] [PubMed] [Google Scholar]
- Quarrie, S. A., S. Pekic Quarrie, R. Radosevic, D. Rancic, A. Kaminska et al., 2006. Dissecting a wheat QTL for yield present in a range of environments: from the QTL to candidate genes. J. Exp. Bot. 57: 2627–2637. [DOI] [PubMed] [Google Scholar]
- Rajaram, S., R. P. Singh and E. Torres, 1988. Current CIMMYT approaches in breeding wheat for rust resistance, pp. 101–118 in Breeding Strategies for Resistance to the Rusts of Wheat, edited by N. W. Simmonds and S. Rajaram. CIMMYT, México, D.F., Mexico.
- Rajaram, S., R. P. Singh and M. VanGinkel, 1996. Approaches to breed for wide adaptation, rust resistance and drought, pp. 2–30 in Proceedings of the 8th Assembly Wheat Breeding Society of Australia, edited by R. A. Richards, C. W. Wrigley, H. M. Rawson, G. J. Rebetzke, J. L. Davidson and R. I. S. Brettell. The Australian National University, Canberra, ACT, Australia.
- Rajaram, S., A. Mujeeb-Kazi and R. P. Singh, 1997. Wheat (Triticum aestivum), pp. 75–144 in Genetics, Cytogenetics and Breeding of Crop Plants, edited by P. N. Bahl, P. M. Salimath and A. K. Mandal. Oxford & IBH Publishing, New Delhi/Calcutta.
- Roelfs, A. P., and D. V. McVey, 1979. Low infection types produced by Puccinia graminis f.sp. tritici. Phytopathology 69: 722–730. [Google Scholar]
- Sawkins, M. C., A. D. Farmer, D. Hoisington, J. Sullivan, A. Tolopko et al., 2004. Comparative map and trait viewer (CMTV): an integrated bioinformatic tool to construct consensus maps and compare QTL and functional genomics data across genomes and experiments. Plant Mol. Biol. 56: 465–480. [DOI] [PubMed] [Google Scholar]
- Sayre, K. D., R. P. Singh, J. Huerta-Espino and S. Rajaram, 1998. Genetic progress in reducing losses to leaf rust in CIMMYT-derived Mexican spring wheat cultivars. Crop Sci. 38: 654–659. [Google Scholar]
- Schnurbusch, T., S. Paillard, A. Schori, M. Messmer, G. Schachermayr et al., 2004. Dissection of quantitative and durable leaf rust resistance in Swiss winter wheat reveals a major resistance QTL in the Lr34 chromosomal region. Theor. Appl. Genet. 108: 477–484. [DOI] [PubMed] [Google Scholar]
- Semagn, K., Å. Bjornstad, H. Skinnes, A. G. Marøy, Y. Tarkegne et al., 2006. Distribution of DArT, AFLP and SSR markers in a genetic linkage map of a double haploid hexaploid wheat population. Genome 49: 545–555. [DOI] [PubMed] [Google Scholar]
- Singh, R. P., 1993. Genetic association of gene Bdv1 for tolerance to barley yellow dwarf virus with genes Lr34 and Yr18 for adult plant resistance to rusts in bread wheat. Plant Dis. 77: 1103–1106. [Google Scholar]
- Singh, R. P., and A. K. Gupta, 1991. Genes for leaf rust resistance in Indian and Pakistani wheats tested with Mexican pathotypes of Puccinia recondita f.sp. tritici. Euphytica 57: 27–36. [Google Scholar]
- Singh, R. P., and R. A. McIntosh, 1984. Complementary genes for reaction to Puccinia recondita tritici in Triticum aestivum. II. Cytogenetic studies. Can. J. Genet. Cytol. 26: 736–742. [Google Scholar]
- Singh, R. P., and S. Rajaram, 1991. Resistance to Puccinia recondita f.sp. tritici in 50 Mexican bread wheat cultivars. Crop Sci. 31: 1472–1479. [Google Scholar]
- Singh, R. P., J. Huerta-Espino, S. Rajaram and J. Crossa, 1998. Agronomic effects from chromosome translocations 7DL.7Ag and 1Bl.1RS in spring wheat. Crop Sci. 38: 27–33. [Google Scholar]
- Singh, R. P., J. C. Nelson and M. E. Sorrells, 2000. Mapping Yr28 and other genes for resistance to stripe rust in wheat. Crop Sci. 40: 1148–1155. [Google Scholar]
- Singh, R. P., J. Huerta-Espino and H. M. William, 2005. Genetics and breeding for durable resistance to leaf and stripe rusts in wheat. Turk. J. Agric. For. 29: 121–127. [Google Scholar]
- Snape, J., M. Foulkes, J. Simmonds, M. Leverington, L. Fish et al., 2007. Dissecting gene × environmental effects on wheat yields via QTL and physiological analysis. Euphytica 154: 401–408. [Google Scholar]
- Smith, A., B. R. Cullis and R. Thompson, 2002. Exploring variety-environment data using random effects models with adjustment for spatial field trends: part 1. Theory, pp.323–335 in Quantitative Genetics, Genomics and Plant Breeding, edited by M. S. Kang. CAB, Wallingford, UK.
- Sorrells, M. E., 2007. Application of new knowledge, technologies, and strategies to wheat improvement. Euphytica 157: 299–306. [Google Scholar]
- Spielmeyer, W., P. J. Sharp and E. S. Lagudah, 2003. Identification and validation of markers linked to a broad-spectrum stem rust resistance gene Sr2 in wheat (Triticum aestivum L.). Crop Sci. 43: 333–336. [Google Scholar]
- Spielmeyer, W., R. A. McIntosh, J. Kolmer and E. S. Lagudah, 2005. Powdery mildew resistance and Lr34/Yr18 genes for durable resistance to leaf and stripe rust cosegregate at a locus on the short arm of chromosome 7D of wheat. Theor. Appl. Genet. 111: 731–735. [DOI] [PubMed] [Google Scholar]
- Suenaga, K., R. P. Singh, J. Huerta-Espino and H. M. William, 2003. Microsatellite markers for Lr34/Yr18 and other quantitative trait loci for leaf rust and stripe rust in bread wheat. Phytopathology 93: 881–890. [DOI] [PubMed] [Google Scholar]
- Thornsberry, J. M., M. M. Goodman, J. Doebley, S. Kresovich, D. Nielson et al., 2001. Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28: 286–289. [DOI] [PubMed] [Google Scholar]
- Trethowan, R., M. P. Reynolds, I. Ortiz-Monasterio and R. Ortiz, 2007. The genetic basis of the Green Revolution in wheat production. Plant Breed. Rev. 28: 39–58. [Google Scholar]
- Van Beem, J., V. Mohler, R. Lukman, M. Van Ginkel, M. William et al., 2005. Analysis of genetic factors influencing the developmental rate of globally important CIMMYT wheat cultivars. Crop Sci. 45: 2113–2119. [Google Scholar]
- van Eeuwijk, F. A., J. B. Denis and M. S. Kang, 1996. Incorporating additional information on genotypes and environments in models for two-way genotype by environment tables, pp.15–49 in Genotype-by-Environment Interaction: New Perspectives, edited by M. S. Kang and H. G. Gauch. CRC Press, Boca Raton, FL.
- Verma, V. A., M. J. Foulkes, A. J. Worland, R. Sylvester-Bradley, P. D. S. Caligari et al., 2004. Mapping quantitative trait loci for flag leaf senescence as a yield determinant in winter wheat under optimal and drought-stressed environments. Euphytica 135: 255–263. [Google Scholar]
- Villareal, R. L., S. Rajaram, A. Mujeeb-Kazi and E. Del Toro, 1991. The effect of chromosome 1B/1R translocation on the yield potential of certain spring wheats (Triticum aestivum L.). Plant Breed. 106: 77–81. [Google Scholar]
- William, H. M., D. Hoisington, R. P. Singh and D. Gonzalez-de-Leon, 1997. Detection of quantitative trait loci associated with leaf rust resistance in bread wheat. Genome 40: 253–260. [DOI] [PubMed] [Google Scholar]
- William, H. M, R. P. Singh, J. Huerta-Espino, S. Ortiz Islas and D. Hoisington, 2003. Molecular marker mapping of leaf rust resistance Lr46 and its association with stripe rust resistance Yr29 in wheat. Phytopathology 93: 153–159. [DOI] [PubMed] [Google Scholar]
- William, H. M., R. P. Singh, J. Huerta-Espino, G. Palacios and K. Suenaga, 2006. Characterization of genetic loci conferring adult plant resistance to leaf rust and stripe rust in spring wheat. Genome 49: 977–990. [DOI] [PubMed] [Google Scholar]
- Worland, A. J., 1996. The influence of flowering time genes on environmental adaptability in European wheats. Euphytica 89: 49–57. [Google Scholar]
- Worland, A. J., A. Börner, V. Korzun, W. M. Li, S. Petrovic et al., 1998. The influence of photoperiod genes on the adaptability on European winter wheats. Euphytica 100: 385–394. [Google Scholar]
- Worland, A. J., E. J. Sayers and V. Korzun, 2001. Allelic variation at the dwarfing gene Rht8 locus and its significance in international breeding programmes. Euphytica 119: 155–159. [Google Scholar]
- Yan, L., A. Loukoainov, G. Tranquilli, M. Helguera, M. Hahima et al., 2003. Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. USA 100: 6263–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., G. Pressoir, W. H. Briggs, I. Vroh Bi, M. Yamasaki et al., 2006. A unified mixed-model method for association mapping that counts for multiple level of relatedness. Nat. Genet. 28(2): 203–208. [DOI] [PubMed] [Google Scholar]
- Xu, X., G. Bai, B. F. Carver, G. E. Shaner and R. M. Hunger, 2005. Molecular characterization of slow leaf-rusting resistance in wheat. Crop Sci. 45: 758–765. [Google Scholar]
- Zhu, Z. D., R. H. Zhou, X. Y. Kong, Y. C. Dong and J. Z. Jia, 2005. Microsatellite markers linked to 2 powdery mildew resistance genes introgressed from Triticum carthlicum accession PS5 into common wheat. Genome 48: 585–590. [DOI] [PubMed] [Google Scholar]