Abstract
Sex is determined in Caenorhabditis elegans through a dose-dependent signal that communicates the number of X chromosomes relative to the ploidy, the number of sets of autosomes. The sex switch gene xol-1 is the direct molecular target of this X:A signal and integrates both X and autosomal components to determine sexual fate. X chromosome number is relayed by X signal elements (XSEs) that act cumulatively to repress xol-1 in XX animals, thereby inducing hermaphrodite fate. Ploidy is relayed by autosomal signal elements (ASEs), which counteract the single dose of XSEs in XO animals to activate xol-1 and induce the male fate. Our goal was to identify and characterize new XSEs and further analyze known XSEs to understand the principles by which a small difference in the concentration of an intracellular signal is amplified to induce dramatically different developmental fates. We identified a new XSE, the ONECUT homeodomain protein CEH-39, and showed that it acts as a dose-dependent repressor of xol-1 transcript levels. Unexpectedly, most other XSEs also repress xol-1 predominantly, but not exclusively, at the transcript level. The twofold difference in X dose between XO and XX animals is translated into the male vs. hermaphrodite fate by the synergistic action of multiple, independent XSEs that render xol-1 active or inactive, primarily through transcriptional regulation.
DURING development, different concentrations of select dose-dependent signals can induce alternative cell fates. Among the classes of dose-dependent signals are those that invoke cell–cell communication to determine developmental fate and those that originate and function within the cell to specify fate. In the first class, signaling molecules secreted from one group of cells influence intracellular signaling cascades in neighboring cells. For example, the Wnt-signaling pathway patterns the dorsal–ventral axis of the Drosophila wing in a concentration-dependent manner. In the second class, gradients of the Drosophila morphogens Bicoid and Nanos exemplify intracellular signals. They control expression of early patterning genes in a concentration-dependent manner to establish anterior–posterior polarity in the embryo (Parisi and Lin 2000; Lynch and Desplan 2003). Defining the molecular nature of dose-dependent signals and their sensors is therefore paramount to understanding cell fate specification in multi-cellular organisms.
Sex-determination strategies reliant on chromosome complement provide further opportunities to dissect mechanisms by which small, quantitative differences in an intracellular signal are translated into alternative developmental fates. For example, in Drosophila melanogaster and Caenorhabditis elegans, chromosome counting mechanisms distinguish one X chromosome from two to specify male (XY or XO) vs. female/hermaphrodite (XX) fate. Both organisms tally the number of X chromosomes relative to the sets of autosomes (Madl and Herman 1979), the X:A ratio, using X-linked genes called X signal elements (XSEs) to communicate the X chromosome number and autosomal signal elements (ASEs) to communicate the ploidy. In D. melanogaster, the double dose of four XSEs (sisA, sisB, sisC, and runt) in diploid XX embryos (X:A = 1.0) activates transcription of the sex switch gene Sex-lethal to induce female development. The single dose of XSEs in diploid XY animals (X:A = 0.5) is insufficient to activate Sex-lethal, thereby permitting the male fate (Cline and Meyer 1996).
In C. elegans, the sex switch gene xol-1 is the direct molecular target of the X:A signal and integrates both X and autosomal components to determine sexual fate. Two copies of XSEs, including the nuclear receptor SEX-1 and the RNA-binding protein FOX-1, induce the hermaphrodite fate in diploid XX embryos by repressing xol-1 through transcriptional and post-transcriptional mechanisms, respectively (Figure 1; Akerib and Meyer 1994; Hodgkin et al. 1994; Nicoll et al. 1997; Carmi et al. 1998). The single copy of XSEs in diploid XO embryos cannot overcome xol-1 activation by the double dose of ASEs, thereby permitting the male fate. One of the ASEs, the T-box transcription factor SEA-1 (signal element on autosome), helps activate xol-1 by increasing its transcript levels (Powell et al. 2005). The X:A signal includes two other partially characterized components, the XSE sex-2 (J. Powell, C. Y. Loh and B. Meyer, unpublished results) and the ASE sea-2 (P. Nix and B. Meyer, unpublished results).
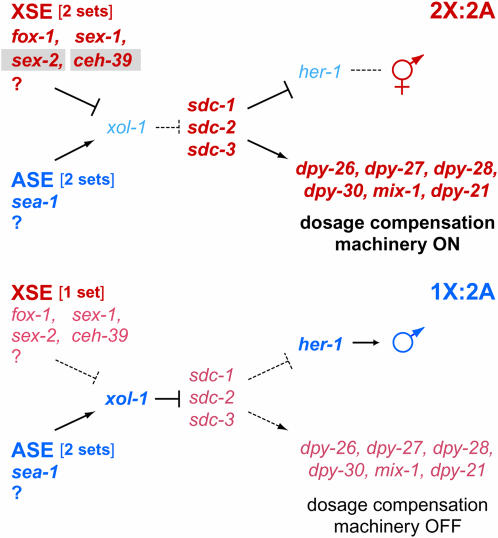
Figure 1.—
The genetic pathway for sex determination and dosage compensation in C. elegans. This pathway includes our discovery of ceh-39 as an XSE and partial analysis of the XSE sex-2; both XSEs are highlighted by gray boxes. In XX animals (top), the two copies of the X-linked XSE genes repress xol-1, permitting activation of the sdc genes. The SDC proteins trigger assembly of the dosage compensation complex (SDC-1, SDC-2, SDC-3, DPY-21, DPY-26, DPY-27, DPY-28, DPY-30, MIX-1) on X, where it reduces gene expression by half. SDC proteins also promote the hermaphrodite fate by repressing her-1, a male sex determination gene. In XO animals (bottom), the single copy of XSEs allows the ASEs to activate xol-1. When xol-1 is active, the sdc genes are repressed, promoting the male fate by permitting her-1 expression and preventing assembly of the dosage compensation machinery on X. The genes in boldface type are active in a specific sex. Genes in red type are required for hermaphrodite development; genes in blue type are required for male development.
The worm sex-determination mechanism discriminates with great accuracy between small differences in the X:A signal. An X:A of 0.67 dictates male fate and an X:A of 0.75 dictates hermaphrodite fate, implying that the effectiveness of the signal might derive from the combined action of multiple X and autosomal elements. Indeed, previous genetic analysis provided evidence that additional X signal elements exist, but did not identify the specific genes (Akerib and Meyer 1994; Carmi and Meyer 1999). Our goal in this study was to identify and characterize new XSEs and to further analyze known XSEs to understand the principles by which intracellular signals can induce different developmental fates in a concentration-dependent manner.
Analysis of the X:A signal is complicated by the fact that the signal controls viability as well as sexual fate. In addition to controlling sex determination, xol-1 controls X chromosome dosage compensation, the vital process that equalizes X-linked gene products between XX and XO animals by halving gene expression from both hermaphrodite X chromosomes (reviewed in Meyer 2005). In XX animals, a decrease in XSE dose or an increase in ASE dose can activate xol-1, prevent dosage compensation, and cause XX lethality. In XO animals, an increase in XSE dose or a decrease in ASE dose can repress xol-1, activate the dosage compensation machinery, and cause XO lethality.
In XO animals, xol-1 sets the male fate by repressing the hermaphrodite-specific sdc genes, which coordinately control downstream genes specialized for regulating either sex determination or dosage compensation (Figure 1). In XX animals, where xol-1 is repressed, SDC-2 induces hermaphrodite development by repressing the male-specific sex-determination gene her-1 and by triggering assembly of the dosage compensation complex (DCC) on both X chromosomes to repress transcript levels. The DCC includes two other SDC proteins and at least seven other dosage compensation proteins, five of which resemble the components of condensin, a conserved protein complex required for mitotic and meiotic chromosome compaction, resolution, and segregation (Villeneuve and Meyer 1987, 1990; Nusbaum and Meyer 1989; Nonet and Meyer 1991; Delong et al. 1993; Chuang et al. 1994; Hsu and Meyer 1994; Lieb et al. 1996, 1998; Davis and Meyer 1997; Kimura and Hirano 1997; Dawes et al. 1999; Hirano 1999; Chu et al. 2002; Yonker and Meyer 2003; M. Albrecht, C. Hassig, C. Tsai and B. Meyer, unpublished results). The DCC binds to recruitment sites on X and then appears to spread in cis to X regions lacking recruitment sites (Csankovszki et al. 2004; McDonel et al. 2006).
Previous studies indicated that the sensitivity and fidelity of X chromosome counting stems from two characteristics: (1) multiple XSEs collaborate to communicate X dose and (2) XSEs use multiple mechanisms to regulate one gene, xol-1. The XSEs act in a cumulative manner to repress xol-1: changing the dose of individual XSEs has little effect on sex determination and dosage compensation, but changing the dose of multiple XSEs has synergistic effects, causing sexual transformation and death (Akerib and Meyer 1994; Carmi and Meyer 1999).
Many principles underlying X chromosome counting have emerged, but a detailed mechanistic picture has not. In our study, we identified the new XSE CEH-39, a ONECUT (OC) homeodomain protein, and further analyzed known XSEs to learn how the X chromosome counting process functions with high precision. Although previous studies showed that both transcriptional and post-transcriptional modes of xol-1 regulation are important, they did not address the relative contribution of each mechanism to xol-1 repression. Our study showed that CEH-39 and most other XSEs communicate X chromosome dose by repressing xol-1 predominately at the transcript level.
MATERIALS AND METHODS
Strains and general methods:
All C. elegans strains were derived from the Bristol variant N2 and were maintained as described in Brenner (1974). Abbreviations are as follows: ceh (C. elegans homeobox), dpy (dumpy), egl (egg-laying defective), fasn (fatty acid synthase), fox (feminizing gene on X), him (high incidence of males), lon (long), nhr (nuclear hormone receptor), sdc (sex determination and dosage compensation), sea (signal element on autosome), sex (signal element on X), tra (sexual transformation), unc (uncoordinated), and xol (XO lethal). The following chromosomal aberrations and mutations were used for this study:
LG II: sea-1(y356) (Powell et al. 2005).
LG III: dpy-27(y57) (Plenefisch et al. 1989), yIs33[Pxol-1∷lacZ] (Nicoll et al. 1997).
LG IV: him-8(e1489), mIs11, yIs2[xol-1∷lacZ] (Rhind et al. 1995), yIs58[ceh-39(+), myo-2∷gfp]. him-8(e1489) increases X chromosome nondisjunction, resulting in 37% XO, 57% XX, and 6% Dpy XXX animals (Hodgkin et al. 1979). mIs11 is a multi-construct array carrying myo-2∷gfp, pes-10∷gfp, and gut∷gfp integrated onto LG IV near dpy-20. yIs58 is an integrated array carrying the wild-type ceh-39 gene and the co-injection marker myo-2∷gfp.
LG X: dpy-3(e27), unc-2(e55), ceh-39(y414), ceh-39(gk296) (Vancouver group of the C. elegans Gene Knockout Consortium), fox-1(y303) (Nicoll et al. 1997), sex-2(y324) (J. Powell and B. Meyer, unpublished results), lon-2(e678), xol-1(y9) (Miller et al. 1988), dpy-6(e14), sex-1(y263) (Carmi et al. 1998).
Duplication: yDp14(X;I) (Akerib and Meyer 1994).
Rearrangement: szT1(I;X) (McKim et al. 1988).
Extrachromosomal array: yEx483[Pdpy-30∷sdc-2(+), myo-2∷gfp(+), rol-6(d)] (Powell et al. 2005).
Mutations not referenced are described in this study or in Riddle et al. (1997).
Isolation of ceh-39(y414):
A C. elegans deletion library was constructed in the Meyer Lab and screened for a ceh-39 deletion following Michael Koelle's C. elegans Gene Knockout Protocol (02/09/03 update) retrieved from his Yale University website (http://info.med.yale.edu/mbb/koelle/). ceh-39 primers used were as follows:
Forward outer primer: GAAATTTACGCTGGCCGTCTGC;
Reverse outer primer: GCCTCTGGATTTCTTTGCTGG;
Forward inner primer: TCTCCGTGCGCTATTTAGGTGC;
Reverse inner primer: TATGGAAGCAGAGCATCGTTGG;
Poison primer 1: CGGTATGTGTTGGAGAAGTCCA;
Poison primer 2: AGAGGTCGTCGACTTCCCAGAG.
RNA interference:
Generally, RNA interference (RNAi) was conducted as described in Kamath et al. (2001), except carbenicillin (25 μg/ml) was used without tetracycline in the overnight cultures. The double-stranded (dsRNA) synthesis was induced in Escherichia coli on plates (1 mm IPTG, 25 μg/ml carbenicillin) incubated overnight at 25°. Bacterial plasmids were constructed or obtained from an Ahringer RNAi feeding library (Kamath and Ahringer 2003). Embryos were placed onto plates with the dsRNA-producing E. coli until they became gravid hermaphrodites (24–36 hr at 20°). Next, two hermaphrodites were picked onto each of six plates with dsRNA-producing E. coli and allowed to lay embryos for 24 hr. The laid embryos were counted, and the resulting animals were scored over a 5-day period to maximize viability estimates for slow-growing worms.
For the matings, males (five per hermaphrodite) were placed onto the original plates containing embryos and dsRNA-producing E. coli once the embryos reached L4. Twenty-four hours later, two gravid, mated hermaphrodites and 10 males were transferred to each of six plates and allowed to mate and lay embryos for 24 hr. The laid embryos were counted. As the animals reached L4, they were picked off and scored. Any animal that failed to reach L4 after 5 days was considered inviable. For progeny counts pertaining to either matings or self-fertilization, the embryos and adults scored for each plate were summed to generate the n values reported in each table, except for strains for which viability was reported with a standard deviation or error of the mean. In those cases, the viability presented is an average of the numbers from the six plates.
For simultaneous RNAi against the three genes ceh-21, ceh-41, and ceh-39, dsRNA corresponding to these genes was injected into the gonads of L4 hermaphrodites. dsRNA was synthesized in vitro with the T7 RiboMAX Large Scale Production System from Promega (Madison, WI) using the Ahringer RNAi feeding construct plasmid DNA as template. dsRNA corresponding to each of these genes was mixed in a 1:1:1 ratio prior to injection. Embryos laid 12–36 hr post-injection were counted, and the resulting adults scored.
Statistical analysis:
Statistical comparisons were made using the χ2 test, except for experiments involving quantitative RT–PCR (qRT–PCR) measurement of transcript levels, which utilized the Student's t-test.
Construction of yIs58:
yIs58 is a UV integrant of the extrachromosomal array yEx689. yEx689 was generated by co-injecting pPD118.33 myo-2∷gfp(+) (50 ng/μl) and pJG75 (50 ng/μl), a plasmid containing a 5.5-kbp genomic PCR fragment spanning the ceh-39 locus amplified with primers (forward, TTTCGGCAAGAGTGCTCTGAAC; reverse, TTGGAATAGAGAAGAGAGCGAC). UV integration involved the following protocol adapted from Andrew Frank. yEx689 worms were washed four times in M9. The worms were then spun down and resuspended in a small volume for plating on an unseeded 9-cm plate. Worms were irradiated without the plate lid using a Stratalinker UV crosslinker (Stratagene, La Jolla, CA) with a UV dose of 15–35 mJ/cm2. OP50 bacteria were then added to the plate, and worms were allowed to recover at room temperature for 5 hr. Transgenic L4 larvae or young adult P0's were plated at a density of two or three per plate on 10–30 plates and allowed to lay F1 progeny, which were then picked individually onto 150 fresh plates. Finally, 2–3 F2 progeny from one F1 plate were picked individually onto 300 fresh plates. Of 300 F2's, 1 segregated 100% GFP-positive animals. The integrated transgene was designated yIs58.
β-Galactosidase staining:
β-Galactosidase activity was used to assess the degree of xol-1 derepression in the reporters yIs2 and yIs33 using X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as the chromogenic substrate for β-galactosidase. yIs2 XX, yIs2; ceh-39(y414) XX, him-8(e1489) yIs2 XX, yIs33 XX, yIs33; him-8(e1489) XX, and yIs33; ceh-39(y414) XX worms were prepared using the following protocol: Worms were placed into a multi-welled glass dish and dried by placing under vacuum for 30 min. Desiccated worms were incubated with −20° acetone for 5 min and allowed to air dry. Worms were then stained by adding staining solution (recipe below) and incubated at 35° for 5–7 hr in a sealed humidified container. The yIs2 and yIs33 him-8 strains were used to control for the time of the β-galactosidase reaction. When the him-8 animals had several darkly stained XO embryos, all reactions were stopped by exchanging the staining solution with H2O. Worms were transferred with a Pasteur pipette to glass slides for microscopy. A worm was considered to have high β-galactosidase activity if it had at least one darkly staining embryo; worms with fewer than three embryos were not scored. The staining solution was prepared from the following: 500 μl 2× phosphate buffer (360 mm Na2HPO4, 40 mm NaH2PO4), 400 μl H2O, 100 μl of 100 mm Redox buffer (50 mm potassium ferricyanide, 50 mm potassium ferrocyanide), 10 μl of 1 m MgCl2, 4 μl 1% SDS, 2 μl of 1 mg/ml 4′,6-diamidino-2-phenylindole (DAPI), 12 μl of 2% X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) in N,N-dimethylformamide, and 5 μl of 50 mg/ml kanamycin sulfate).
Quantification of transcript levels:
qRT–PCR was used to measure transcript levels from RNA isolated from three independent growths of the strains listed in Table 7. The protocol of Van Gilst et al. (2005) was used, except that worms were grown on egg plates (http://www.wormbook.org) prior to isolating the mixed-stage embryos, and the total RNA was treated with DNase prior to cDNA synthesis using 3 μl of RQ1 RNase-free DNase (Promega)/100 μg of RNA in a 50-μl reaction, as per manufacturer's instructions. For each strain tested, 5 μg of total RNA were used to generate cDNA. Primer sequences are available upon request.
TABLE 7.
XSE mutations increase xol-1 transcript levels
| Transcript measuredby qRT–PCRa
|
||
|---|---|---|
| Genotype | xol-1 | nhr-64 |
| sex-1(y263) | 3.0 ± 0.5 | 1.1 ± 0.1 |
| sex-2(y324) | 1.8 ± 0.2 | 1.2 ± 0.1 |
| ceh-39(y414) | 2.0 ± 0.2 | 1.1 ± 0.1 |
| ceh-39(gk296) | 2.0 ± 0.3 | 1.1 ± 0.1 |
| fox-1(y303) | 1.0 ± 0.1 | 1.3 ± 0.1 |
The levels of xol-1 and XSE transcripts in mutant embryos (listed by genotype) were measured by qRT–PCR and are expressed as the fold change compared to the transcript levels measured in wild-type embryos (see materials and methods). For example, a value of 2.0 means that twice as many gene-specific transcripts were measured in mutant embryos than in wild-type embryos. All transcript levels were normalized to the levels of the control gene, fasn-1 (ORF F32H2.5), whose expression is constant throughout embryogenesis and is not affected by dosage compensation. See Van Gilst et al. (2005) for details and protocol. nhr-64, another gene not affected by dosage compensation, was used as a control to gauge the variability and reliability of measurements made using qRT–PCR. Experimental error is expressed as the standard error of the mean. A critical control was to compare the fasn-1-normalized xol-1 or nhr-64 transcript levels in three independent preparations of wild-type embryos. That comparison showed the xol-1 and nhr-64 transcript levels to be statistically equivalent among the independent RNA preparations (xol-1, 1.2 ± 0.2; nhr-64, 1.3 ± 0.1).
Transcript levels of genes indicated in Table 7 were normalized to the transcript level of the fatty acid synthase gene fasn-1 [open reading frame (ORF) F32H2.5], which is expressed constitutively throughout embryogenesis, by adjusting the cycle threshold (Ct) value of fasn-1, measured in each strain to equal the Ct value of fasn-1 measured in wild-type animals. The Ct values of all other transcripts measured in the same strain were then adjusted by the same amount. This adjustment equalizes the small variations in concentration of the starting material added to each PCR reaction from different RNA preparations.
The transcript level of each mutant strain was then expressed as fold change relative to wild-type animals (ΔCt). The normalized Ct value for each transcript measured in each strain was subtracted from the normalized Ct value of the same transcript measured in wild-type animals. The difference between these values corresponds to the change in transcript levels relative to those in wild-type animals. Ct values are expressed as PCR cycle numbers. Each PCR cycle increases the concentration of the template by twofold. Therefore, to convert the difference in Ct values to a relative change in concentration, the expression 2ΔCt was used.
CEH-39 antibody:
Two separate rabbit anti-CEH-39 antibodies (CA1184 and CA1183) were raised (Covance) against a 28-amino-acid peptide including the CEH-39 N terminus plus a GC linker (DFSNTYRNYGEVVDFPEDFESDYVPTVKGC). Both antibodies were affinity purified using the same peptide, which was synthesized by David King (University of California, Berkeley). Both antibodies yielded similar staining patterns. For neither antibody was staining detectable in mutants carrying the ceh-39(y414) deletion, which eliminates the DNA encoding the peptide. CA1184 was used for Westerns and embryo staining (Figure 4, A–D). CA1183 was used for gonad staining (Figure 4, E and F).
Figure 4.—
CEH-39 accumulates in nuclei of young embryos, consistent with its role as an XSE, and also in germline nuclei. (A–D) Partial projections of false-colored confocal images of wild-type and ceh-39 mutant embryos stained with DAPI (red) and antibodies (CA1184) against CEH-39 (green). (A and B) CEH-39 localizes in a diffuse pattern within interphase nuclei of young (50-cell) and older (150- to 200-cell) XX embryos; CEH-39 also associates with mitotic chromosomes (arrow and inset in A). (C) CEH-39 is greatly reduced in embryos with >200 cells. (D) No CEH-39 antibody staining was detectable in ceh-39 deletion mutant embryos, which lack the antibody epitope, demonstrating specificity of the CEH-39 antibody. Bars, 10 μm. (E and F) Partial projections of false-colored confocal images of wild-type and ceh-39 mutant gonads stained with DAPI (red) and CEH-39 antibodies (green) (CA1183). Two focal planes (separated by a dashed white line in E) were used to show pachytene and diplotene diakinesis. In late pachytene and early diplotene, CEH-39 staining appears diffuse nuclear and excluded from the nucleolus. In late diplotene and diakinesis, staining colocalizes with condensed chromosomes. Enlargement of the nucleus in diakinesis is shown in insets in E. Staining is absent in gonads of ceh-39 deletion mutants.
Immunofluorescence microscopy:
Embryos were fixed as described in Davis and Meyer (1997) and stained as described in Chuang et al. (1994), except that both the primary and the secondary antibody staining were done overnight. The following antibodies were used: rabbit anti-CEH-39, rabbit anti-DPY-27 (Chuang et al. 1994), rat anti-SDC-3 (McDonel et al. 2006), FITC-conjugated goat anti-rabbit (Jackson ImmunoResearch Labs), and Cy5-conjugated donkey anti-rabbit (Jackson ImmunoResearch Labs). Fixed and stained embryos were mounted in VectaShield (Vector Laboratories, Burlingame, CA) containing 0.5 μg/ml of DNA intercalating dye DAPI. At least 1000 embryos were examined for each experiment. Gonads were fixed and stained as in Howe et al. (2001). All images were captured on a Leica TCS NT microscope. Images of all embryos or gonads in Figures 3 and 4 are projections of four 0.5-μm sections.
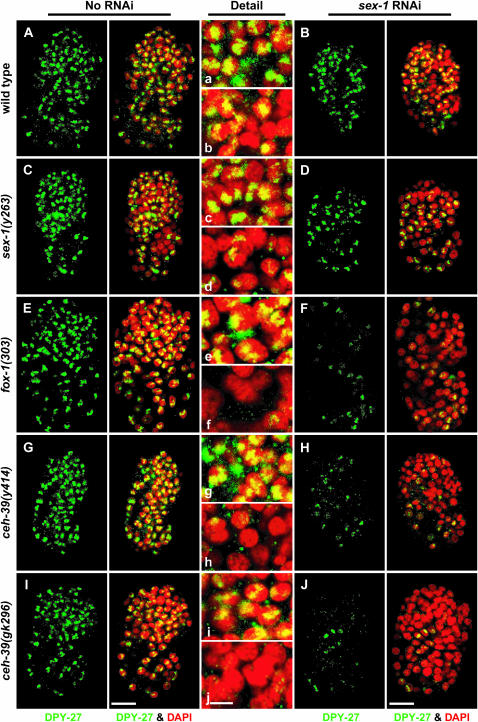
Figure 3.—
Decreasing the dose of ceh-39 and other XSEs disrupts the dosage compensation complex in XX animals. Localization of DPY-27 in wild-type and XSE mutant embryos with and without RNAi disruption of sex-1. (A–J) Partial projections of false-colored confocal images of wild-type and mutant XX embryos costained with antibodies against DPY-27 (green) and the DNA intercalating dye DAPI (red). (a–j) Enlargements of nuclei from A–J, respectively. (A, E, G, and I) DPY-27 localized in a punctate pattern to the X chromosomes of wild-type XX embryos, and fox-1 or ceh-39 mutant XX embryos. (C) The sex-1(y263) mutant embryos exhibited a reduction in DPY-27 staining. Some nuclei showed sparse or no staining and others varied from punctate to diffuse staining, all implying less DPY-27 on X. (B and D) sex-1(RNAi) XX embryos and sex-1(y263, RNAi) mutants showed a further decrease in DPY-27 signal compared to that in sex-1 mutants. The residual DPY-27 was mostly punctate, indicating X localization. (F, H, and J) The DPY-27 signal was drastically reduced in fox-1 sex-1(RNAi) XX and ceh-39 sex-1(RNAi) XX mutants; the small quantity of residual DPY-27 had punctate localization. The more severe reduction in DPY-27 signal after knockdown of two XSEs rather than one shows that dosage compensation is disrupted more in double than in single XSE mutants, as is viability. In the images, DPY-27 signal was enhanced in all embryos treated with RNAi of sex-1 to demonstrate the punctate localization more clearly. Bars: A–J, 10 μm; a–j, 3 μm.
RESULTS
Identification of the X signal element ceh-39:
Previous analysis of duplications and deficiencies at the left end of X defined three distinct regions that harbor X signal elements, but only the XSE in region 3 (fox-1) was discovered (Figure 2A; Akerib and Meyer 1994; Hodgkin et al. 1994; Nicoll et al. 1997; Carmi and Meyer 1999). We designed an RNAi-based screen to identify ORFs in region 2 that function as XSEs (Figure 2, A and B). All 146 region 2 ORFs were assayed for XSE activity utilizing the sensitized strain yDp14/yDp14 (X;I); him-8 IV; fox-1 X, in which 94% of XO animals die from the increased dose of XSEs. The fox-1 mutation sensitizes the screen to permit identification of weak XSEs. The homozygous yDp14 duplication triples the dose of fox-1, the XSEs in region 2, and other potential XSEs adjacent to regions 2 and 3 (Akerib and Meyer 1994), causing XO animals to die from inappropriate repression of xol-1 and the consequent reduction of X-linked gene expression (Table 1A). In principle, reducing the cumulative XSE activity in this strain by RNAi disruption of an XSE gene should increase the proportion of viable XO males, thus forming the basis for an efficient and sensitive assay to screen for XSE activity. An RNAi screen is more advantageous than a genetic screen, because RNAi reduces the activity of all copies of an XSE, whereas a mutation reduces only the activity of the single copy on X or on the duplication. Therefore, XSEs with even minor contributions to the signal should emerge from this screen. Our approach was validated by the observation that RNAi-mediated reduction of fox-1 activity increased viability of yDp14/yDp14; fox-1 males from 6 to 84% (Table 1A).
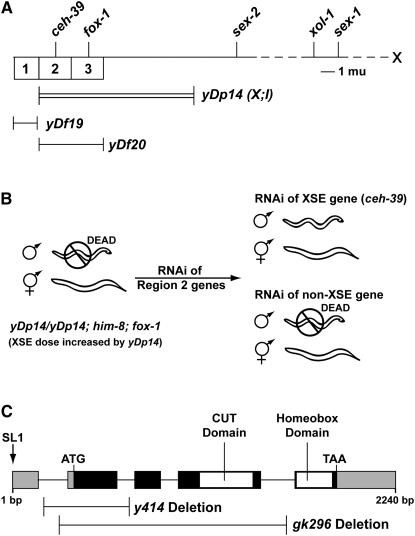
Figure 2.—
Genetic map of the X chromosome, the RNAi-based screen for identifying XSEs in region 2, and the genomic region of ceh-39. (A) The X map highlights XSEs and xol-1 (above the line) and three regions (numbered boxes) shown previously using duplications and deficiencies to contain X signal elements. The duplication yDp14 covers region 2 (ceh-39) and region 3 (fox-1), the deficiency yDf19 uncovers region 1, and the deficiency yDf20 uncovers regions 2 and 3. (B) Screen for XSEs in region 2. The homozygous duplication yDp14 increases XSE dose sufficiently in XO animals to repress xol-1, causing complete XO lethality. To identify potential XSEs, each ORF in region 2 was targeted for RNAi in yDp14/yDp14; him-8; fox-1 animals, and their progeny were scored for the presence of males. Of all genes tested, only RNAi of ceh-39 suppressed XO lethality, indicating that ceh-39 is a potential XSE in region 2. (C) The genomic region spanning the ceh-39 locus. Exons are indicated by solid boxes. The cut and homeobox domains are indicated by open boxes in the exons; 5′- and 3′-UTRs are indicated by shaded boxes. The arrow indicates where the SL1 trans-spliced leader is spliced to the 5′-UTR. ATG and TAA are the translational start and stop codons, respectively. The genomic region of ceh-39 is 2240 bp, including introns, which are indicated by lines between the boxes. Locations of the two deletions in the ceh-39 locus, y414 and gk296, are indicated by single lines.
TABLE 1.
ceh-39 is an XSE in region 2
| yDp14/yDp14; him-8; fox-1 XO + RNAi of genea | Male viability (%)b | nc |
|---|---|---|
| A. RNAi of ceh-39 suppresses the XO-specific lethality caused by the increase in XSE dose from two copies of yDp14 | ||
| No RNAi vector or gene | 6 | 1225 |
| RNAi vector with no gene | 18d | 1236 |
| fox-1(RNAi) | 84d | 1050 |
| ceh-39(RNAi) | 84d | 857 |
| ceh-21(RNAi) | 20d | 776 |
| ceh-41(RNAi) | 28d | 923 |
| ceh-39(RNAi), ceh-21(RNAi), ceh-41(RNAi) | 76e | 946 |
| RNAi of X ORFs not in region 2 | 19 ± 9f (13 genes) | NA |
| RNAi of ORFs on autosomes | 18 ± 7f (14 genes) | NA |
| XO genotypeg | ceh-39 and fox-1 dose | Male viability (%) | nc |
| B. ceh-39 and fox-1 are not the only XSEs in yDp14 | |||
| Wild typeh | 1 | 100 | 1632 |
| yDp14/+i | 2 | 61 | 724 |
| yDp14/+; ceh-39(y414) fox-1(y303)j | 1 | 98 | 1369 |
| yDp14/+; ceh-39(gk296) fox-1(y303)j | 1 | 102 | 1566 |
| yDp14/yDp14; him-8(e1489)b | 3 | 0 | 929 |
| yDp14/yDp14; ceh-39(y414) fox-1(y303)k | 2 | 8 | 863 |
| yDp14/yDp14; ceh-39(gk296) fox-1(y303)k | 2 | 1 | 716 |
Candidate genes were tested for XSE activity. yDp14/yDp14(X;I); him-8(e1489) IV; dpy-3(e27) fox-1(y303) unc-2(e55) X hermaphrodites were treated with RNAi against the indicated gene, and the viability of progeny males was assessed. In all cases except as described in footnote e, the RNAi was achieved through feeding. RNAi-mediated knockdown of an XSE should decrease the male lethality caused by the increase in XSE dose from yDp14. Animals were fed bacteria that produced dsRNA to the listed gene (see materials and methods). yDp14 is an X duplication attached to LG I and can exist in one copy (yDp14/+) or two copies (yDp14/yDp14) (Akerib and Meyer 1994). him-8 XX animals produce 37% XO males, 57% XX hermaphrodites, and 6% Dpy XXX hermaphrodites (Hodgkin et al. 1979).
Male viability was calculated by the following formula: (no. of adult males)/(expected no. of males) × 100. The number of expected males was (0.37)n.
n is the total number of embryos from six independent sets of progeny counts.
Male viability is significantly higher only for fox-1(RNAi) and ceh-39(RNAi), both P ≤ 0.01, when compared to male viability of the true control: yDp14/yDp14; him-8; fox-1 animals grown on bacteria carrying an RNAi vector with no gene insert. Male viability for neither ceh-21(RNAi) (P = 0.70) nor ceh-41(RNAi) (P = 0.02) was significantly different from the true control. Male viability due to RNAi of these genes instead was equivalent to that due to RNAi of random X ORFs not in region 2 or of autosomal ORFs. The unexpected observation that RNAi against any C. elegans gene, or even the introduction of double-stranded RNA not similar to C. elegans RNA, rescued some XO lethality caused by yDp14/yDp14 suggests that the RNAi machinery may affect the sex-determination and dosage compensation pathway. This RNAi effect appears to be weak since it was observed only in the sensitized XO genetic background and only when RNAi was achieved through dsRNA feeding. Induction of RNAi in XX animals did not cause a notable dosage compensation disruption, and thus the observed effects of RNAi in XX animals (Table 3) are due to the reduced function of the XSE genes targeted.
RNAi was achieved by simultaneously injecting double-stranded RNA from ceh-39, ceh-21, and ceh-41. Injection RNAi against fox-1 resulted in 87% male viability (n = 1217); against ceh-39, 85% male viability (n = 759); and against dsRNA made from the vector with no cloned gene, 6% male viability (n = 1002).
The numbers presented include the average and the standard deviation of male viability for RNAi against 13 X ORFs not in region 2 (C05D9.5, F49E7.1, C05D9.7, R193.2, R193.3, R193.1, T13G4.3, F09E10.3, F09E10.6, F09E10.7, F09E10.8, K06A9.1, K06A9.2) and 14 ORFs on autosomes (F44E8.2, C31H1.1, C31H1.2, C31H1.5, C31H1.6, C31H1.7, C31H1.8, C10G6.1, T10B9.3, T10B9.4, T10B9.5, T10B9.7, T10B9.8, ZK938.1). Approximately 200 embryos were scored per ORF tested.
These animals also carry a dpy-3(e27) mutation, except for yDp14/+ and yDp14/yDp14; him-8(e1489) animals, which carry unc-2(e55) instead.
Males were generated by mating wild-type males and hermaphrodites. Male viability was calculated by the following formula: [adult males]/[expected no. of males, (0.5)n] × 100. The number of hermaphrodites was 0.5(n), implying a viability of 100% and a mating that produced only cross progeny.
Males were produced by mating wild-type males with yDp14/yDp14; unc-2(e55) hermaphrodites. The number of hermaphrodites was (0.5)n, indicating that the hermaphrodite viability was 100% and the cross went to completion.
yDp14/+; ceh-39 fox-1 males were generated from a cross of mIs11 males with yDp14/yDp14; ceh-39 fox-1 hermaphrodites. mIs11 is a dominant, integrated transgenic marker that expresses GFP from pes-10 and myo-2 promoters and a gut-specific enhancer. It was used to identify cross progeny. Male viability was calculated by the following formula: [adult males)]/[expected no. of males, (0.5)n] × 100. All progeny were gfp(+), indicating that the cross went to completion.
yDp14/yDp14; mIs11/+; ceh-39(y414 or gk296) fox-1(y303) males were generated by crossing yDp14/+; mIs11; ceh-39(y414 or gk296) fox-1(y303) males with yDp14/yDp14; ceh-39(y414 or gk296) fox-1(y303) hermaphrodites. Fifty percent of the XO (male) cross progeny should be of genotype yDp14/yDp14; ceh-39(y414 or gk296) fox-1(y303). Since another 50% of the XO progeny are yDp14/+; ceh-39(y414 or gk296) fox-1(y303), which are ∼100% viable, the viability of yDp14/yDp14; ceh-39(y414 or gk296) fox-1(y303) XO males was calculated by the following formula: [no. of males − (0.25)n]/[expected no. of yDp14/+ males, (0.25)n] × 100.
Of all 146 ORFs in region 2, only RNAi disruption of the gene corresponding to the ORF called T26C11.7 increased male viability significantly (P ≤ 0.01), enhancing it to 84% and suggesting that T26C11.7 is an XSE (Table 1A). On average, RNAi against 13 random X ORFs not in region 2 or against 14 random ORFs on autosomes enhanced male viability to ∼20%, a value not significantly different from the viability of yDp14/yDp14; fox-1 males grown on bacteria containing the RNAi vector lacking a candidate gene (Table 1A). That the introduction of any dsRNA into the yDp14/yDp14; fox-1 XO animals enhanced viability to this extent suggests that the RNAi machinery might affect dosage compensation, a topic currently under investigation (see Table 1, footnote d).
T26C11.7 represents the gene ceh-39, which encodes a member of the OC class of homeodomain proteins. The DNA-binding domain (DBD) of OC proteins is characterized by an atypical homeobox domain and a single cut domain. OC proteins mediate transcriptional regulation of numerous developmental processes (Lannoy et al. 1998). The discovery that an XSE in region 2 encodes a putative transcription factor came as somewhat of a surprise, since previous work suggested that region 2 likely contained a post-transcriptional regulator of xol-1 (Nicoll et al. 1997). Experiments described below confirm that ceh-39 regulates xol-1 transcript levels and reconcile previous results.
C. elegans encodes five additional OC proteins, and the genes for two, ceh-21 and ceh-41, reside on X in an operon with ceh-39 (Blumenthal et al. 2002; Burglin and Cassata 2002). However, neither ceh-21 nor ceh-41 behaves like an XSE: RNAi against ceh-21 or ceh-41 increased the viability of yDp14/yDp14; fox-1 XO animals to only 20 or 28%, respectively, levels comparable to the average levels achieved by RNAi against ORFs on autosomes or X chromosome ORFs not in region 2 (Table 1A). Moreover, simultaneous RNAi disruption of ceh-21, ceh-41, and ceh-39 did not further increase the viability of yDp14/yDp14; fox-1 XO animals (Table 1A). Thus, XSE activity is specifically a property of ceh-39 and not of other X-linked OC genes.
To characterize ceh-39 genetically, two ceh-39 mutants were isolated (Figure 2C; see materials and methods). ceh-39(y414) deletes part of the ceh-39 locus, resulting in a conceptual protein that lacks the first 102 amino acids but retains both the homeobox and cut domains. The allele ceh-39(gk296) is a larger deletion that eliminates the N terminus and the cut domain. XX and XO animals carrying either mutation have a wild-type phenotype (Table 3 and data not shown). Both mutations synergize with a fox-1 mutation to suppress all the male lethality caused by one copy of yDp14, providing genetic confirmation that the ceh-39 locus behaves like an XSE (Table 1B).
TABLE 3.
ceh-39 mutations enhance XX-specific phenotypes caused by other XSE mutations
| Genotypea | Survivor phenotype | Hermaphrodite viability (%)b | nc |
|---|---|---|---|
| ceh-39(y414) | Wild type | 101 | 1008 |
| ceh-39(gk296) | Wild type | 102 | 1021 |
| ceh-39(RNAi) | Wild type | 100 | 807 |
| ceh-21(RNAi) | Wild type | 100 | 1147 |
| fox-1(y303) | Wild type | 99 | 1054 |
| sex-2(y324) | Wild type to mild Dpy | 99 | 1032 |
| sex-1(y263) | Dpy, Egl Tra | 70 | 884 |
| sex-1(RNAi) | Very Dpy, Egl, Tra | 17 | 1090 |
| sex-1(y263, RNAi) | Very Dpy, Egl, Tra | 17 | 1304 |
| sex-2(y324) sex-1(y263)d | Very Dpy | 4 | 238 |
| fox-1(y303) sex-1(y263)e | Very Dpy, Egl, Tra | 4 | 1176 |
| fox-1(y303) sex-1(RNAi) | Very Dpy, Egl, Tra | 9 | 749 |
| ceh-39(y414) sex-1(y263)f | Very Dpy, Egl, Tra | 7 | 982 |
| ceh-39(y414) sex-1(y263)e | Very Dpy, Egl, Tra | 5 | 941 |
| ceh-39(RNAi) sex-1(y263) | Very Dpy, Egl | 10 | 832 |
| ceh-39(y414) sex-1(RNAi) | Dead | 0 | 935 |
| ceh-39(gk296) sex-1(RNAi) | Dead | 0 | 1637 |
| ceh-21(RNAi) sex-1(y263) | Dpy, Egl | 75 | 1043 |
| ceh-39(y414) fox-1(RNAi) | Mild Dpy | 98 | 1486 |
| ceh-39(gk296) fox-1(RNAi) | Mild Dpy | 101 | 1256 |
| ceh-39(RNAi) fox-1(y303) | Mild Dpy | 100 | 729 |
| ceh-39(RNAi) sex-2(y324) | Dpy, Egl | 99 ± 2g | 1368 |
| fox-1(y303) sex-2(y324) | Dpy, Egl | 98 ± 1g | 1010 |
| ceh-39(RNAi) fox-1(y303) sex-2(y324) | Dpy, Egl | 92 ± 1g | 1180 |
RNAi was applied as explained in Table 1, footnote a.
Hermaphrodite viability was calculated by the following formula: (no. of adult hermaphrodites)/(total no. of embryos) × 100.
n is the total number of embryos from six independent sets of progeny counts.
Data are from C. Y. Loh and B. J. Meyer (personal communication). Of 951 progeny from sex-2(y324) sex-1(y263)/szT1 animals, only 9 (of an expected 238) lacked szT1, implying that they were sex-2(y324) sex-1(y263) and only 4% were viable. sex-2(y324) sex-1(y263) animals were severely Dpy and produced no or few progeny.
Percentage viability of fox-1(y303) sex-1(y263) XX progeny from the strain fox-1(y303) sex-1(y263)/szT1 and percentage viability of ceh-39(y414) sex-1(y263) XX progeny from the strain ceh-39(y414) sex-1(y263)/szT1 were calculated by the following formula: (no. of Dpy hermaphrodites)/0.25(n − no. of males) × 100. The szT1 balancer acts as a mild dominant him mutation, making it necessary to calculate the expected number of XX adults by subtracting the number of XO male progeny from the total number of embryos.
This strain is maintained under yEx483[dpy-30∷sdc-2(+); myo-2∷gfp(+)], an extrachromosomal array that rescues XSE mutants because it overexpresses sdc-2. To score viability, progeny from gfp(−) hermaphrodites that had lost yEx483 were counted.
Viability was calculated separately for six independent sets of progeny counts. Average viability and error are reported. Error is expressed as the standard error of the mean.
ceh-39 and fox-1 are not the only XSEs in the yDp14 interval:
If ceh-39 and fox-1 were the only significant contributors to the cumulative XSE dose in yDp14, then the ceh-39 fox-1 double mutations should rescue all yDp14/+ males and restore the viability of yDp14/yDp14 males to that of yDp14/+ males. All yDp14/+; ceh-39(y414 or gk296) fox-1 males appeared viable (Table 1B). However, the viability of yDp14/yDp14; ceh-39 fox-1 XO animals was only ∼2–13% of the viability of yDp14/+ XO animals. Thus, ceh-39 and fox-1 are important XSEs in the interval of X represented by yDp14, but they are not the only XSEs. This duplicated region extends beyond region 3 by 7 MU (including >450 ORFs), suggesting that additional XSEs reside in this region of X.
ceh-39 acts upstream of xol-1:
If ceh-39 is a bona fide XSE, it should exert its effect on dosage compensation by repressing xol-1, rather than a downstream gene in the dosage compensation pathway. In XX animals, mutations in XSEs derepress xol-1, causing disruption of dosage compensation and the consequent XX-specific phenotypes, including lethality, an egg-laying defect (Egl), and dumpy (Dpy) morphology, all of which are suppressed by a xol-1 mutation (Akerib and Meyer 1994; Carmi et al. 1998). Although ceh-39 mutations by themselves cause no obvious dosage compensation phenotype in XX mutants, they enhance the XX-specific lethality caused by hypomorphic mutations in dosage compensation genes such as dpy-27 or sdc-2, which act downstream of xol-1 (Figure 1). If ceh-39 acts through xol-1, then a xol-1 mutation should suppress the synergistic XX lethality caused by the combination of dpy-27 and ceh-39 mutations or ceh-39 and sdc-2 mutations. Moreover, the triple-mutant XX animals (dpy-27; ceh-39 xol-1 or ceh-39 xol-1 sdc-2) should have the same phenotypes as either dpy-27 or sdc-2 single mutants, respectively.
The hypomorphic dpy-27(y57) mutation reduced XX viability to 77% (Table 2; Plenefisch et al. 1989), and the dpy-27; ceh-39(y414) double combination further reduced XX viability to 18%; survivors had more severe Dpy and Egl phenotypes. The synergistic XX lethality was almost completely suppressed by a xol-1 null mutation: ∼70% of dpy-27; ceh-39 xol-1 XX animals were viable, indicating that ceh-39 functions upstream of xol-1 to repress it (Table 2). The synergistic lethality between ceh-39(y414) and sdc-2(RNAi) was also suppressed by a xol-1 mutation (Table 2), further confirming that ceh-39 controls xol-1 either directly or indirectly.
TABLE 2.
ceh-39 acts upstream of xol-1
| Genotypea | Hermaphrodite viability (%)b | nc |
|---|---|---|
| ceh-39(y414) | 101 | 1008 |
| xol-1(y9)d | 97 | 1251 |
| ceh-39(y414) xol-1(y9)d | 93 | 1120 |
| dpy-27(y57) | 77 | 1164 |
| dpy-27(y57); xol-1(y9) | 77 | 1435 |
| dpy-27(y57); ceh-39(y414) | 18 | 1130 |
| dpy-27(y57); xol-1(y9) ceh-39(y414)d | 70 | 964 |
| sdc-2(RNAi) | 84 | 1512 |
| xol-1(y9) sdc-2(RNAi)d | 84 | 722 |
| ceh-39(y414) sdc-2(RNAi) | 53 | 1157 |
| ceh-39(y414) xol-1(y9) sdc-2(RNAi)d | 89 | 1416 |
RNAi was applied as explained in Table 1, footnote a.
Hermaphrodite viability was calculated by the following formula: (no. of adult hermaphrodites)/(total no. of embryos) × 100.
n is the total number of embryos from six independent sets of progeny counts.
Strain also includes the marker dpy-6(e14).
Criteria for an X signal element:
The hallmark of an X signal element is that changing its dose causes reciprocal effects on the viability of XX and XO animals. First, decreasing XSE dose selectively kills XX animals by activating xol-1 and thereby inhibiting the dosage compensation machinery. Second, increasing XSE dose selectively kills XO animals by repressing xol-1 and thereby activating the dosage compensation machinery. XSEs act cumulatively such that increasing or decreasing the dose of multiple XSEs affects viability more severely than changing the dose of a single XSE. Third, increasing the dose of one XSE in XX animals compensates for decreasing the dose of a different XSE, and decreasing the dose of one XSE in XO animals compensates for increasing the dose of another. The compensating changes restore the cumulative XSE signal to a level approaching that of the wild-type signal. Fourth, an XSE acts in a dose-dependent manner in the zygote, since the zygotic X:A signal determines sex. Results described in the sections below show that ceh-39 meets these criteria and therefore acts as an X signal element in promoting the hermaphrodite fate.
Decreasing ceh-39 dose enhances the XX lethality caused by reduced XSE dose:
Mutations in individual XSEs have small-to-moderate effects on XX animals, but mutations in multiple XSEs can cause pronounced dosage compensation defects and extensive XX-specific lethality (Table 3; Carmi and Meyer 1999). For example, 70% of hypomorphic sex-1 XX mutants are viable, and virtually all fox-1 XX or sex-2 XX single mutants are viable, but nearly all fox-1 sex-1 or sex-2 sex-1 XX double mutants are dead (P < 0.01; Table 3); Carmi et al. 1998). Similarly, decreasing ceh-39 dose enhances the XX lethality caused by loss of other XSEs. While neither ceh-39 RNAi nor a ceh-39 mutation causes visible phenotypes, both cause nearly complete XX lethality (10 and 7%, respectively) in combination with a sex-1 hypomorphic mutation (P < 0.01; Table 3). This effect is XX specific: all ceh-39 sex-1 XO double mutants are viable (data not shown). In contrast, RNAi of ceh-21 did not enhance the lethality of sex-1 XX mutants (Table 3), demonstrating that synergistic lethality is not a general property of OC gene disruptions and that the general process of RNAi does not demonstrably affect the X signal in XX animals.
ceh-39 and fox-1 are relatively weak XSEs, but even weak XSEs make important contributions to the X signal (Table 3). ceh-39 XX, fox-1 XX, or sex-2 XX mutants are wild type. However, ceh-39 fox-1 XX double mutants are mildly Dpy in phenotype (Table 3), as are ceh-39(RNAi) sex-2 XX double mutants and fox-1 sex-2 XX double mutants, suggesting that ceh-39 and fox-1 make similar contributions to the X signal. The triple-mutant combination ceh-39 fox-1 sex-2 caused slightly more Dpy and Egl phenotypes than double-mutant combinations and a slight reduction in viability (92% viable, P < 0.01) compared to double mutants (98–100% viable), further illustrating the cumulative action of XSEs.
Decreasing the dose of ceh-39 and other XSEs disrupts the DCC in XX animals:
The extent of XX-specific lethality caused by mutations in one or more XSEs was found to be well correlated with the degree of disruption in the dosage compensation complex (Figure 3, A–J). In wild-type XX animals, the dosage compensation complex assembles on both X chromosomes to reduce transcript levels by half (Chuang et al. 1994; Lieb et al. 1996, 1998; Davis and Meyer 1997; Dawes et al. 1999; Chu et al. 2002). Consistent with the full viability and wild-type appearance of fox-1 XX or ceh-39 XX mutants, the DCC proteins DPY-27 (Figure 3, A and a; E and e; G and g; I and i) and SDC-3 (data not shown) exhibited a wild-type X-localized pattern. The DCC was only mildly disrupted in sex-1 hypomorphic mutants (70% viable) (Figure 3, C and c) and more severely disrupted in sex-1(RNAi) XX animals (Figure 3, B and b) or sex-1(y263, RNAi) XX mutants (Figure 3, D and d), both of which were ∼17% viable. In the latter two cases, DPY-27 and SDC-3 (data not shown) appeared punctate in many nuclei and diffuse or absent in others. The most dramatic disruption of the DCC was evident in fox-1 sex-1(RNAi) embryos (Figure 3, F and f) and ceh-39 sex-1(RNAi) embryos (Figure 3, H and h; J and j), all of which were inviable. Most nuclei had very little or no DPY-27 or SDC-3 (data not shown) protein, and the residual protein had either a diffuse nuclear or punctate appearance. This dramatic reduction in DCC levels suggests that the complete lethality observed in these double mutants is due to a disruption in dosage compensation.
Increasing ceh-39 dose enhances the XO lethality caused by increased XSE dose:
Increasing the dose of ceh-39 using yIs58[ceh-39(+)], an integrated array bearing multiple copies of the 5.5-kbp genomic fragment spanning ceh-39, caused no XO lethality by itself but enhanced the lethality caused by one copy of yDp14, which duplicates ceh-39, fox-1, and other not-yet-identified XSEs (Table 4). One copy of yIs58[ceh-39(+)] reduced the viability of yDp14/+ XO males from 61 to 20% (P < 0.01). Two copies of yIs58[ceh-39(+)], shown to increase the ceh-39 transcript level fourfold above that of the wild-type level (Gladden et al. 2007, accompanying article in this issue), further reduced viability of yDp14/+ males to 3%, suggesting that ceh-39 represses xol-1 in a dose-dependent manner, another characteristic of XSEs (P < 0.01 for viability of yIs58/+ vs. yIs58/yIs58 XO animals).
TABLE 4.
Increased dose of ceh-39 enhances XO-specific lethality caused by increased XSE dose
| XO genotypes | No. of males | Male viability (%)a | nb |
|---|---|---|---|
| Wild typec | 812 | 100 | 1632 |
| yIs58[ceh-39(+)]/yIs58d | 555 | 98 | 1131 |
| yDp14/+e | 220 | 61 | 724 |
| yDp14/+; yIs58[ceh-39(+)]/+f | 141 | 20 | 1440 |
| yDp14/+; yIs58[ceh-39(+)]/yIs58g | 22 | 3 | 1522 |
Males were generated through crosses and their viability was calculated by the following formula: [no. of adult males]/[the expected no. of males, (0.5)n] × 100. In all crosses, the number of hermaphrodites was 0.5(n), implying that the matings produced only cross progeny and that hermaphrodite viability was 100%.
n is the total number of embryos from six independent sets of progeny counts.
Wild-type males were produced by mating wild-type males and hermaphrodites.
Males were produced by mating yIs58[ceh-39(+)]/yIs58 males and hermaphrodites. yIs58[ceh-39(+)] is an integrated transgenic array carrying multiple copies of a 5.5-kbp genomic fragment spanning the entire ceh-39 locus. Two copies of yIs58 elevate the ceh-39 transcript level fourfold above the wild-type level (Gladden et al. 2007, accompanying article in this issue).
Males were produced by mating wild-type males with yDp14/yDp14; unc-2(e55) hermaphrodites.
Males were generated by mating yIs58[ceh-39(+)]/yIs58 males with yDp14/yDp14; unc-2(e55) hermaphrodites.
Males were generated by mating yIs58[ceh-39(+)]/yIs58 males with yDp14/yDp14; yIs58/yIs58; unc-2(e55) hermaphrodites.
Changes in ceh-39 dose compensate for reciprocal changes in the dose of other XSEs:
The screen used to identify ceh-39 demonstrated that decreasing the dose of ceh-39 suppressed the XO-specific lethality caused by elevated XSE dose in yDp14/yDp14; fox-1 animals (Table 1). Reciprocal suppression also occurred: increasing ceh-39 dose suppressed the XX-specific lethality caused by all combinations of single and double XSE mutations tested (Table 5). Two copies of yIs58[ceh-39(+)] increased the viability of sex-1 XX mutants from 70 to 98%, fox-1 sex-1 mutants from 0 to 76%, and sex-2 sex-1 mutants from 4 to 69% (P < 0.01 for all pairwise comparisons). The rescued XX mutants were Dpy, indicating lingering dosage compensation defects despite high viability. This rescue was specific to ceh-39, since ceh-39 RNAi not only abolished the ability of yIs58[ceh-39(+)] to rescue XSE mutants, but also actually caused synergistic lethality in single and double XSE mutants (Table 5). Thus, increasing ceh-39 dose can compensate for a decreased dose of other XSEs, thereby restoring the X signal and repressing xol-1.
TABLE 5.
Overexpression of ceh-39 rescues XX-specific lethality caused by disruption of other XSE genes
| Genotypea | Hermaphrodite viability (%)b | nc |
|---|---|---|
| sex-1(y263) | 70 | 884 |
| sex-1(y263); yIs58[ceh-39(+)]/yIs58[ceh-39(+)]d | 98 | 1399 |
| sex-1(y263, RNAi) | 17 | 1304 |
| sex-1(y263, RNAi); yIs58/yIs58 | 71 | 597 |
| fox-1(y303) sex-1(y263) | 4 | 1176 |
| fox-1(y303) sex-1(y263); yIs58/yIs58 | 76 | 784 |
| sex-2(y324) sex-1(y263)e | 4 | 238 |
| sex-2(y324) sex-1(y263); yIs58/yIs58 | 69 | 1286 |
| ceh-39(RNAi) sex-1(y263); yIs58/yIs58f | 5 | 440 |
| ceh-39(RNAi) fox-1(y303) sex-1(y263); yIs58/yIs58f | 0 | 480 |
RNAi was applied as explained in Table 1, footnote a.
Hermaphrodite viability was calculated by the following formula: (no. of adult hermaphrodites)/(total no. of embryos) × 100.
n is the total number of embryos from six independent sets of progeny counts.
yIs58[ceh-39(+)] is an integrated transgene consisting of multiple copies of a 5.5-kbp genomic fragment spanning the ceh-39 locus.
Data are from C. Y. Loh and B. J. Meyer (personal communication). See footnote d in Table 3.
The rescue of XSE mutants is specific to increased ceh-39 dose because RNAi of ceh-39 not only abolished the suppression caused by yIs58, but also reduced the activity of ceh-39 completely, causing synergistic lethality in combination with XSE mutations.
ceh-39 acts in the zygote:
Sex is specified by the X chromosome dose of the zygote. Thus, for ceh-39 to be classified as an XSE, changing the dose of ceh-39 in the zygote should perturb viability. In the previously described experiments showing that increasing ceh-39 dose adversely affected XO animals, the increased ceh-39 dose from one copy of yIs58[ceh-39(+)] in yDp14/+ XO males was supplied paternally, indicating that the increase in XO lethality resulted from a change in zygotic activity of ceh-39. Thus, ceh-39 meets the fourth and final criterion for an XSE.
ceh-39 does not regulate known XSEs or the ASE sea-1:
The results presented thus far show that ceh-39 acts as an XSE to repress xol-1. ceh-39 could function by activating another XSE by repressing an activator of xol-1 such as an ASE or by acting directly on xol-1. If ceh-39 activates a single known XSE, then mutation of both should cause the same degree of xol-1 derepression as loss of just the downstream XSE, provided the downstream XSE mutation is a null. Instead, loss of ceh-39 activity causes synergistic XX-specific dosage compensation phenotypes in combination with null mutations (or RNAi) of XSEs, making it unlikely that ceh-39 acts through them (Table 3). Reinforcing this conclusion is the fact that increasing the ceh-39 dose rescues all known XSE mutants (Table 5).
If ceh-39 represses a specific ASE, then mutation of ceh-39 should increase expression of the target ASE gene and thereby hyperactivate xol-1 in an XX embryo, causing a dosage compensation disruption. Mutation of the target ASE would suppress the ceh-39 mutation by preventing the upregulation of xol-1 expression. This scenario was not found for the ASE sea-1. sea-1 activates xol-1 expression, and a sea-1 mutation rescues 57% of fox-1 sex-1 mutants by reducing xol-1 activation (Table 6; Powell et al. 2005). If ceh-39 were a repressor of sea-1, then a sea-1 mutation would block the complete synergistic XX lethality between ceh-39 RNAi and fox-1 sex-1 mutations. Instead, if ceh-39 acts independently of sea-1, then ceh-39 RNAi should reduce the viability of fox-1 sex-1 mutants even in the presence of a sea-1 mutation. Indeed, ceh-39 RNAi did reduce the viability of sea-1; fox-1 sex-1 XX mutants from 57 to 0% (P < 0.01; Table 6). Therefore, ceh-39 appears not to act through sea-1.
TABLE 6.
An autosomal signal element mutation cannot suppress loss of three XSEs
| Genotypea | Hermaphrodite viability (%)b | nc |
|---|---|---|
| sea-1(y356); fox-1(y303) sex-1(y263) | 57 | 1054 |
| sea-1(y356); fox-1(y303) sex-1(y263) ceh-39(RNAi) | 0 | 2016 |
RNAi was applied as explained in Table 1, footnote a.
Hermaphrodite viability was calculated by the following formula: (no. of adult hermaphrodites)/(total no. of embryos) × 100.
n is the total number of embryos from six independent sets of progeny counts.
Furthermore, ceh-39 is unlikely to be the exclusive regulator of any ASE. If ceh-39 were the sole and complete repressor of a single ASE, the ceh-39 dose should be sufficiently high in XX animals to repress the ASE. If this were the case, increasing ceh-39 dose in XX animals should not alleviate xol-1 derepression caused by mutations in other XSEs, since its target ASE would already be repressed. However, increasing ceh-39 dose strongly suppresses single and double XSE mutants (Table 5). Together, the results suggest that ceh-39 acts as a direct repressor of xol-1 or functions as an activator of an undefined XSE.
CEH-39 accumulates in a spatial and temporal pattern appropriate for an XSE:
The expression patterns of known XSEs match the time window in which xol-1 repression is critical: during gastrulation from the ∼28- to 350-cell stage (Rhind et al. 1995; Nicoll et al. 1997; Carmi et al. 1998). To assess whether CEH-39 accumulation is consistent with its role as an XSE, we raised an antibody against CEH-39 and examined its immuno-localization in wild-type and ceh-39 mutant animals. In wild-type embryos, CEH-39 was first detectable in the 2-cell stage, but robust CEH-39 accumulation began at the 8-cell stage and tapered off by the 150-cell stage, disappearing almost completely by the 200-cell stage (Figure 4, A–C), consistent with a role in repressing xol-1. No antibody staining was detected in ceh-39 mutants, confirming antibody specificity (Figure 4D). CEH-39 also appeared to associate with condensed DNA. During mitosis, CEH-39 was detected on metaphase and anaphase chromosomes (Figure 4A). However, no obvious mitotic defects were found in ceh-39 mutants, suggesting that the accumulation of this OC protein on condensed DNA may simply reflect a nonspecific affinity for DNA or a minor function in mitosis. The presence of CEH-39 in 2-cell embryos and also in hermaphrodite gonads (Figure 4, E and F) correlates with a previous study showing that the XSE in region 2, the location of ceh-39, has a maternal component (Carmi and Meyer 1999). In the gonad, CEH-39 nuclear staining was observed from late pachytene through diakinesis. Staining colocalized with the condensed diakinetic chromosomes.
ceh-39 requires sequences in the xol-1 coding region to repress it:
The identification of CEH-39 as a OC transcription factor suggested that CEH-39 would regulate xol-1 transcript levels. However, previous studies suggested that the XSE in region 2 did not act on a transcriptional level (Carmi et al. 1998). In XX animals, a heterozygous deficiency that uncovers region 2 and fox-1 (yDf20) failed to derepress a xol-1 transcriptional Pxol-1∷lacZ reporter transgene (yIs33), in which lacZ expression was controlled by the 2.8-kbp xol-1 promoter. In contrast, both a sex-1 mutation and a heterozygous deficiency that uncovers region 1 (yDf19) caused robust derepression of yIs33 (Figure 2A; Figure 5, A and D; Nicoll et al. 1997; Carmi et al. 1998). Thus, fox-1 and the XSE in region 2 appeared to repress xol-1 post-transcriptionally, while the XSE in region 1 and sex-1 appeared to repress xol-1 transcriptionally. This interpretation was reinforced by finding that yIs33 was also not derepressed in the ceh-39(y414) deletion mutant (Figure 5D). However, these results did not preclude the possibility that ceh-39 might regulate xol-1 transcript levels through sites not present in the yIs33 reporter.
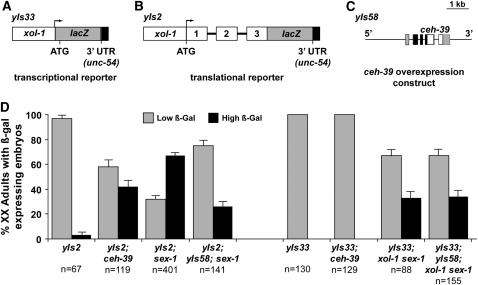
Figure 5.—
ceh-39 represses xol-1 expression via genomic sequences spanning the first three exons of xol-1. (A) yIs33 is an integrated xol-1 transcriptional reporter transgene containing the xol-1 promoter (2.8 kbp) (open box) fused in-frame to the lacZ gene (shaded box) and the unc-54 3′-UTR (solid box). (B) yIs2 is an integrated xol-1 translational reporter transgene containing the xol-1 promoter (2.8 kbp) and genomic sequences spanning the first three exons of xol-1 fused in-frame with the lacZ gene (shaded box) and the unc-54 3′-UTR (solid box). The promoter and exons are represented by open boxes and introns by lines. Both y1s2 and yIs33 recapitulate the regulation of xol-1: high expression in XO embryos and low expression in XX embryos. (C) yIs58 is an integrated ceh-39-overexpressing transgene containing a 5.5-kbp genomic fragment spanning the ceh-39 promoter (2.3 kbp), coding region, and 1 kbp of downstream sequence. (D) β-Galactosidase levels were qualitatively estimated and binned for sex-1, ceh-39, and yIs58; sex-1 mutant animals carrying either yIs2 or yIs33. A xol-1 mutation was included in strains with yIs33 and a sex-1 mutation to suppress the synergistic XX lethality. XX embryos were considered to have high levels of β-galactosidase activity if the intensity of staining matched that of XO animals and low levels of β-galactosidase activity if the staining ranged from none to less intense than that of XO animals. Both sex-1 and ceh-39 mutants derepress yIs2, as assessed by the increased percentages of adults bearing embryos strongly expressing β-galactosidase; sex-1 and ceh-39 must repress xol-1 through sequences present in yIs2. Overexpressing CEH-39 via yIs58 reduced the percentage of sex-1 mutant adults expressing β-galactosidase, indicating that increased levels of CEH-39 can restore repression to xol-1 in the absence of sex-1 through xol-1 genomic sequences spanning the first three exons. In contrast, ceh-39 mutants did not derepress the xol-1 transcriptional reporter yIs33, unlike sex-1 mutants. Consistent with this finding, overexpressing CEH-39 via yIs58 did not restore xol-1 repression in sex-1 mutants, indicating that ceh-39 does repress xol-1 through its promoter sequences. Error bars represent the standard deviation of a binomial distribution.
To examine the regulation of xol-1 by ceh-39 more extensively, a reporter transgene encompassing a larger genomic region of xol-1 was analyzed. Results from these experiments indicate that ceh-39 represses xol-1 through sequences present in the xol-1-coding region. In this reporter, the lacZ gene is under the control of a genomic fragment spanning the 2.8-kbp xol-1 promoter and the first three exons (yIs2, xol-1∷lacZ) (Figure 5B). Both sex-1, and to a lesser extent, ceh-39 mutations derepress yIs2. Homozygous sex-1(y263) and ceh-39(y414) mutations caused 67 and 42%, respectively, of yIs2 XX hermaphrodites to produce embryos expressing high levels of lacZ compared to only 3% of yIs2 XX control hermaphrodites (P < 0.01; Figure 5D). These results show that loss of ceh-39 alone is sufficient to derepress xol-1. The fact that the ceh-39 mutation causes less derepression of yIs2 than the sex-1 mutation suggests that ceh-39 is a weaker repressor of xol-1 than sex-1, consistent with the weaker phenotypes of ceh-39 mutants.
Because ceh-39 is a xol-1 repressor, increasing the ceh-39 dose should counteract the derepression of xol-1 caused by the loss of another XSE such as sex-1. In fact, increasing ceh-39 dose using the integrated transgene yIs58[ceh-39(+)] reduced the extent of yIs2 derepression caused by a sex-1 mutation (Figure 5, C and D). Only 26% of yIs2; yIs58/yIs58; sex-1(y263) XX hermaphrodites produced embryos expressing high levels of lacZ, in contrast to 67% of yIs2; sex-1(y263) hermaphrodites (P < 0.01). Furthermore, the increase in ceh-39 dose had no effect on the derepression of yIs33 by sex-1 mutations, consistent with CEH-39 acting through xol-1 sequences not in the promoter. That is, an equivalent number of hermaphrodites produced embryos expressing high levels of lacZ in both the yIs33; sex-1(y263) strain and the yIs33; yIs58/yIs58; sex-1(y263) strain (Figure 5D). These results show ceh-39 to be a repressor of xol-1 that acts independently of sex-1. The function of CEH-39, but not that of SEX-1, depends on sequences spanning the first three exons of xol-1.
ceh-39 and other XSEs reduce xol-1 transcript levels:
The changes observed in the expression of xol-1 reporters show that ceh-39 represses xol-1 but do not establish whether ceh-39 regulates xol-1 transcript levels. Therefore, qRT–PCR was used to measure the total level of xol-1 transcripts in both wild-type and XSE mutant XX embryos using xol-1 primer sets designed to measure all splice variants of xol-1 simultaneously (Table 7).
Mutations that inactivate transcriptional repressors of xol-1 should increase xol-1 transcripts, but mutations that disrupt post-transcriptional regulators should not. Our data show that ceh-39, sex-1, and sex-2 regulate total xol-1 transcript levels, but fox-1 does not (Table 7).
xol-1 expression is 10 times lower in XX than in XO embryos (Rhind et al. 1995). Since XSEs act cumulatively, mutations in individual XSEs would be expected to increase xol-1 transcript levels <10-fold compared to wild-type levels. In fact, a sex-1 mutation caused an increase in xol-1 transcript levels 3-fold above wild-type levels (P < 0.01), consistent with a role as a transcriptional repressor, while a fox-1 mutation had no obvious effect on xol-1 transcript levels, as expected from its role as a post-transcriptional regulator (Table 7). Both deletion alleles of ceh-39 increased xol-1 transcript levels ∼2-fold above wild-type levels in XX embryos (P < 0.01, Table 7). Together with xol-1 reporter data, these results indicate that ceh-39 regulates xol-1 transcript levels either directly through a regulatory element present in the genomic sequence spanning the first three exons of xol-1 or indirectly by unknown means.
The XSE sex-2 also affects the total level of xol-1 transcripts. A sex-2 mutation, like a ceh-39 mutation, increased xol-1 transcript levels approximately twofold above wild-type levels in XX embryos (P < 0.01; Table 7). These results show that four of five XSEs affect xol-1 transcript levels. Repression of xol-1 transcript levels appears to be the most prevalent mode of xol-1 regulation by XSEs; however, post-transcriptional repression by XSEs such as fox-1 plays an important role in xol-1 regulation (Figure 6; Tables 3, 5, and 7; Nicoll et al. 1997; Carmi and Meyer 1999).
Figure 6.—
Regulation of xol-1 through XSEs and ASEs. xol-1 is the direct molecular target of the X:A signal and integrates both X and autosomal components to determine sexual fate. The molecular diagram indicates where the XSEs act to repress xol-1 and where the ASEs function to activate xol-1. Our study showed that CEH-39 and most other XSEs communicate X chromosome dose by repressing xol-1 transcript levels. The XSE in region 1 and SEX-1 (nuclear hormone receptor) repress xol-1 through sequences in the promoter, while CEH-39 (homeodomain protein) acts through xol-1 genomic sequences that span the first three exons. CEH-39 may also function through promoter sequences. The means by which sex-2 represses xol-1 transcript levels has not been defined. Using a separate mechanism, FOX-1, an RNA-binding protein, represses xol-1 on a post-transcriptional level. Both the transcriptional and post-transcriptional mechanism are important for xol-1 repression. The ASE SEA-1 (T-box protein) activates xol-1 transcript levels using promoter sequences. XSEs and ASEs could compete directly to regulate xol-1 by binding overlapping or neighboring cis regulatory sites or indirectly by affecting components of the transcriptional machinery. The end result is that the higher XSE activity in XX animals out-competes ASE activity and inactivates xol-1, but the lower XSE activity in XO animals permits the ASE activity to activate xol-1. The high level of XOL-1 protein present in XO animals then induces the male fate, including repression of the dosage compensation machinery. The lower level of XOL-1 in XX animals permits the hermaphrodite fate, including activation of the dosage compensation machinery.
DISCUSSION
We addressed the fundamental question of how a small difference in the concentration of an intracellular signal is amplified to induce different cell fates. In C. elegans, sex is determined through a dose-dependent signal that translates the twofold difference in X chromosome dose between XO and XX diploid embryos into the male or hermaphrodite fate by switching the xol-1 sex-determination gene on or off. The sex signal, the X:A ratio, consists of autosomal signal elements that activate xol-1 and of X signal elements that repress it. In this study, we identified the X signal element CEH-39, a ONECUT homeodomain transcription factor. CEH-39 functions in the sex-determination pathway upstream of xol-1 to communicate X chromosome dose by repressing xol-1 transcript levels in a dose-dependent manner. Furthermore, we showed that four of the five known XSEs control xol-1 at this level of regulation, suggesting that the sensitivity of the sex-determination signal stems in part from synergistic interactions among multiple repressors acting at the transcript level.
Function of the ONECUT homeodomain protein CEH-39 in determining sex:
OC homeodomain proteins are a conserved class of transcription factors that normally contain a bipartite DNA-binding domain composed of a single cut domain and an atypical homeodomain (Lemaigre et al. 1996; Burglin and Cassata 2002). OC proteins can stimulate transcription by recruiting the CREB-binding protein coactivator through an LXXLL motif in the cut domain (Lannoy et al. 2000). The OC DBD can also function as a coactivator with the Forkhead box family of transcription factors, which contain winged-helix DBDs (Rausa et al. 2003). Although most OC proteins act as transcriptional activators, at least one example exists in which an OC protein acts to inhibit transcription rather than stimulate it, by antagonizing the activity of another transcription factor (Pierreux et al. 1999).
Here we have demonstrated the function of an OC protein as a dose-dependent repressor. The DBD of CEH-39 is similar to other OC proteins in organisms as diverse as insects and mammals, yet outside the DBD, CEH-39 bears no similarity to other OC proteins, including CEH-21 and CEH-41, two C. elegans OC proteins encoded in an operon with CEH-39 (Burglin and Cassata 2002). Both CEH-21 and CEH-41 contain the conserved sequence element OCAM (ONECUT-associated motif) that CEH-39 lacks, and ceh-41 lacks the cut domain in its DBD. Functional analysis showed that, of the three coregulated OC genes, only ceh-39 functions as an XSE. Disruption of neither ceh-21 nor ceh-41 had any affect on sex determination or dosage compensation in the sensitized XX and XO genetic backgrounds used.
CEH-39's action as a dose-dependent repressor of xol-1 transcript levels requires a 350-bp region of xol-1 spanning its first three exons, thus identifying new sites of xol-1 important for the control of its transcript levels. Previous analysis of SEX-1 revealed that transcriptional regulation of xol-1 occurred through its promoter (Carmi et al. 1998). Similarly, other xol-1 regions might also be necessary for regulation by CEH-39, but they are not sufficient to confer repression without this 350-bp region. Thus, in its capacity as a transcriptional repressor, CEH-39 could, in principle, bind not only to cis regulatory elements outside the xol-1 promoter, but also to ones within the promoter. The 350-bp region contains two core consensus binding sites (ATCAAT) established for the mammalian OC protein HNF-6 (Lannoy et al. 1998). The C. elegans OC protein CEH-21 binds to DNA sequences containing this consensus site (Lannoy et al. 1998), indicating that OC-binding specificity is evolutionarily conserved. Since both ceh-39 and ceh-21 encode bipartite OC DBDs, it is likely that CEH-39 also binds sequences bearing ATCAAT. These consensus binding sites reside in the xol-1 promoter as well as within the first three exons, providing the opportunity for cooperation in the binding of CEH-39 molecules to distant sites within the xol-1 locus. Indeed, preliminary electromobility shift assays using the recombinant CEH-39 protein suggest that CEH-39 binds to multiple, OC consensus-like sites in the xol-1 promoter and in exon 3.
Function of the X signal:
Our work has shown that transcriptional regulation is the predominant but not exclusive form of xol-1 repression by the multi-genic X signal (Figure 6). Previous analysis had predicted that the XSE in region 2 would regulate xol-1 at the post-transcriptional level and that transcriptional and post-transcriptional regulation might be equally important (Nicoll et al. 1997). The discovery of the CEH-39 transcriptional regulator in region 2 and the observation that sex-2 also controls xol-1 transcript levels suggests that more XSEs control xol-1 at the transcriptional level rather than at the post-transcriptional level. However, at least one XSE, the RNA-binding protein FOX-1, is a potent post-transcriptional regulator of xol-1, indicating that multiple levels of regulation function to relay the X signal to the sex determination and dosage compensation machinery. The combined action of two separate mechanisms enhances xol-1 repression.
Studies of multi-component nucleoprotein complexes, called enhanceosomes and repressosomes, which activate or repress transcription in eukaryotes, may provide a molecular explanation for the phenotypic synergy observed among XSEs. Cooperative binding of multiple independent activators or repressors to each other and to DNA leads to synergistic changes in transcription by, for example, altering the recruitment of the basal RNA polymerase II transcriptional machinery to a promoter (Carey 1998; Ptashne and Gann 2002; Gowri et al. 2003; Ptashne 2004). The result of this synergy is that small changes in the concentration of multiple regulators causes a greater-than-additive transcriptional response of their target gene. The fact that XSEs function synergistically and that at least four XSEs (ceh-39, sex-1, sex-2, and the XSE in region 1) affect xol-1 transcript levels opens the possibility that these XSEs function as part of a repressosome recruited to xol-1 regulatory regions.
Our genetic evidence indicates that ceh-39 functions both independently of sex-1 and synergistically with it to repress xol-1. These two XSEs require different sites in the xol-1 locus to regulate it. SEX-1, a nuclear hormone receptor, represses xol-1 transcription through its binding sites in the xol-1 promoter (Carmi et al. 1998). CEH-39 requires xol-1 sequences spanning the first three exons to repress it, although OC consensus binding motifs occur in both the promoter and the exons, suggesting that CEH-39 could actually bind to either or both locations. Thus, the possibility exists for CEH-39 and SEX-1 to bind xol-1 in a cooperative manner. However, the fact that ceh-39 and sex-1 may potentially bind nonadjacent sites in the xol-1 locus suggests that if they are part of a repressosome, (1) their binding may change DNA structure to bring the sites into close proximity or (2) the two XSEs may collaborate indirectly through interactions with the general transcriptional machinery.
XSEs vs. ASEs:
The question arises as to why so many XSEs regulate xol-1 transcript levels. The recent characterization of ASEs provides a clue. Loss of the two known ASEs (sea-1 and sea-2) reduces xol-1 transcript levels, suggesting that a significant component of the autosomal signal activates xol-1 through transcriptional mechanisms (Powell et al. 2005; P. Nix and B. J. Meyer, unpublished results). With both the X and autosomal components of the primary sex signal regulating xol-1 at the level of transcription, these opposing elements could compete for the control of the regulatory machinery that sets xol-1 expression levels. XSEs and ASEs could compete directly by binding overlapping or neighboring cis regulatory targets or indirectly by influencing components of the transcriptional machinery. Direct competition could endow XSEs and ASEs with the dose sensitivity necessary to assess X chromosome number. The signal with the higher activity could out-compete the opposing signal and gain control of xol-1 expression. In XX animals, the high XSE activity would inactivate xol-1, but in XO animals, the lower XSE activity would permit the relatively higher ASE activity to turn on xol-1.
Relative strength of XSEs:
XSEs function cumulatively to repress xol-1, and an individual XSE appears to contribute a relatively small portion of the X signal, thus accounting, in part, for the large number of XSEs (at least five). However, not all XSEs contribute equally to the X signal; their relative strengths differ significantly. sex-1 appears to make the strongest individual contribution: loss of sex-1 activity causes substantial XX-specific lethality, unlike the loss of any other single XSE. In fact, sex-1 appears stronger than ceh-39, fox-1, and sex-2 combined. Paradoxically, a sex-1 mutation derepresses xol-1 transcript levels by only 1.5-fold more than an XSE mutation that causes insignificant XX lethality. Furthermore, even though ceh-39 overexpression rescues the lethality of sex-1 mutants and reduces xol-1 transcript levels to that of ceh-39 mutants (Figure 5), which have no overt phenotype, the ceh-39-overexpressing sex-1 mutants are still somewhat Dpy. How can that be? The fact that these mutants are still Dpy despite low xol-1 transcript levels suggests that sex-1's role in the sex-determination pathway may be multi-faceted. sex-1 may affect the sex-determination pathway independently of xol-1. If sex-1 has a dual role as an XSE and a xol-1 independent regulator of sex determination and dosage compensation, then mutations in sex-1 (1) would cause phenotypes more severe than expected on the basis of its effects on xol-1 expression and (2) would not be completely suppressed by increasing the dose of another XSE. Both appear to be true, and further analysis of sex-1 in the accompanying article in this issue (Gladden et al. 2007) establishes the involvement of sex-1 in other aspects of sex determination and dosage compensation.
In summary, our analysis of the dose-dependent sex-determination signal in C. elegans revealed important insights into the strategy by which the twofold difference in X dose between XO and XX animals is translated into the male vs. hermaphrodite fate by rendering xol-1 active or inactive. We identified a new component of the X signal, ceh-39, and showed that the dose dependence of the sex-determination signal derives in part from the regulation of xol-1 transcript levels by multiple, independent XSEs.
Acknowledgments
We thank M. Van Gilst for guidance with quantitative RT–PCR, D. King for designing and producing the CEH-39 peptide for antibody production, P. Nix for providing strains and information about ASEs, J. Powell and C. Y. Loh for strains and information about sex-2, the C. elegans Gene Knockout Consortium for providing the ceh-39(gk296) mutant, T. Cline for discussions, and I. Carmi, B. Farboud, M. Jow, and A. Severson for critical comments on the manuscript. This work was supported by National Institutes of Heath grant GM30702 to B.J.M. and by a National Science Foundation Predoctoral Fellowship to J.M.G. B.J.M. is an investigator of the Howard Hughes Medical Institute.
References
- Akerib, C. C., and B. J. Meyer, 1994. Identification of X chromosome regions in Caenorhabditis elegans that contain sex-determination signal elements. Genetics 138: 1105–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal, T., D. Evans, C. D. Link, A. Giffanti, D. Lawson et al., 2002. A global analysis of Caenorhabditis elegans operons. Nature 417: 851–854. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burglin, T. R., and G. Cassata, 2002. Loss and gain of domains during evolution of cut superclass homeobox genes. Int. J. Dev. Biol. 46: 115–123. [PubMed] [Google Scholar]
- Carey, M., 1998. The enhanceosome and transcriptional synergy. Cell 92: 5–8. [DOI] [PubMed] [Google Scholar]
- Carmi, I., and B. J. Meyer, 1999. The primary sex determination signal of Caenorhabditis elegans. Genetics 152: 999–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmi, I., J. B. Kopczynski and B. J. Meyer, 1998. The nuclear hormone receptor SEX-1 is an X-chromosome signal that determines nematode sex. Nature 396: 168–173. [DOI] [PubMed] [Google Scholar]
- Chu, D. S., H. E. Dawes, J. D. Lieb, R. C. Chan, A. F. Kuo et al., 2002. A molecular link between gene-specific and chromosome-wide transcriptional repression. Genes Dev. 16: 796–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang, P. T., D. G. Albertson and B. J. Meyer, 1994. DPY-27: a chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell 79: 459–474. [DOI] [PubMed] [Google Scholar]
- Cline, T. W., and B. J. Meyer, 1996. Vive la différence: males vs. females in flies vs. worms. Annu. Rev. Genet. 30: 637–701. [DOI] [PubMed] [Google Scholar]
- Csankovszki, G., P. McDonel and B. J. Meyer, 2004. Recruitment and spreading of the C. elegans dosage compensation complex along X chromosomes. Science 303: 1182–1185. [DOI] [PubMed] [Google Scholar]
- Davis, T. L., and B. J. Meyer, 1997. SDC-3 coordinates the assembly of a dosage compensation complex on the nematode X chromosome. Development 124: 1019–1031. [DOI] [PubMed] [Google Scholar]
- Dawes, H. E., D. S. Berlin, D. M. Lapidus, C. Nusbaum, T. L. Davis et al., 1999. Dosage compensation proteins targeted to X chromosomes by a determinant of hermaphrodite fate. Science 284: 1800–1804. [DOI] [PubMed] [Google Scholar]
- Delong, L., J. D. Plenefisch, R. D. Klein and B. J. Meyer, 1993. Feedback control of sex determination by dosage compensation revealed through Caenorhabditis elegans sdc-3 mutations. Genetics 133: 875–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladden, J. M., B. Farboud and B. J. Meyer, 2007. Revisiting the X:A signal that specifies Caenorhabditis elegans sexual fate. Genetics 177: 1639–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowri, P. M., J. H. Yu, A. Shaufl, M. A. Sperling and R. K. Menon, 2003. Recruitment of a repressosome complex at the growth hormone receptor promoter and its potential role in diabetic nephropathy. Mol. Cell. Biol. 23: 815–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, T., 1999. SMC-mediated chromosome mechanics: A conserved scheme from bacteria to vertebrates? Genes. Dev. 13: 11–19. [DOI] [PubMed] [Google Scholar]
- Hodgkin, J., H. R. Horvitz and S. Brenner, 1979. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91: 67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., J. D. Zellan and D. G. Albertson, 1994. Identification of a candidate primary sex determination locus, fox-1, on the X chromosome of Caenorhabditis elegans. Development 120: 3681–3689. [DOI] [PubMed] [Google Scholar]
- Howe, M., K. L. McDonald, D. G. Albertson and B. J. Meyer, 2001. HIM-10 is required for kinetochore structure and function on Caenorhabditis elegans holocentric chromosomes. J. Cell Biol. 153: 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, D. R., and B. J. Meyer, 1994. The dpy-30 gene encodes an essential component of the Caenorhabditis elegans dosage compensation machinery. Genetics 137: 999–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R. S., and J. H. Ahringer, 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 4: 313–321. [DOI] [PubMed] [Google Scholar]
- Kamath, R. S., M. Martinez-Campos, P. Zipperlen, A. G. Fraser and J. Ahringer, 2001. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2: RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, K., and T. Hirano, 1997. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell 90: 625–634. [DOI] [PubMed] [Google Scholar]
- Lannoy, V. J., T. R. Burglin, G. G. Rousseau and F. P. Lemaigre, 1998. Isoforms of hepatocyte nuclear factor-6 differ in DNA-binding properties, contain a bifunctional homeodomain, and define the new ONECUT class of homeodomain proteins. J. Biol. Chem. 273: 13552–13562. [DOI] [PubMed] [Google Scholar]
- Lannoy, V. J., A. Rodolosse, C. E. Pierreux, G. G. Rousseau and F. P. Lemaigre, 2000. Transcriptional stimulation by hepatocyte nuclear factor-6 target-specific recruitment of either CREB-binding protein (CBP) or p300/CBP-associated factor (p/CAF). J. Biol. Chem. 275: 22098–22103. [DOI] [PubMed] [Google Scholar]
- Lemaigre, F. P., S. M. Durviaux, O. Truong, V. J. Lannoy, J. J. Hsuan et al., 1996. Hepatocyte nuclear factor 6, a transcription factor that contains a novel type of homeodomain and a single cut domain. Proc. Natl. Acad. Sci. USA 93: 9460–9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb, J. D., E. E. Capowski, P. Meneeley and B. J. Meyer, 1996. DPY-26, a link between dosage compensation and meiotic chromosome segregation in the nematode. Science 274: 1732–1736. [DOI] [PubMed] [Google Scholar]
- Lieb, J. D., M. R. Albrecht, P. T. Chuang and B. J. Meyer, 1998. MIX-1: an essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell 92: 265–277. [DOI] [PubMed] [Google Scholar]
- Lynch, J., and C. Desplan, 2003. ‘De-evolution’ of Drosophila toward a more generic mode of axis patterning. Int. J. Dev. Biol. 47: 497–503. [PubMed] [Google Scholar]
- Madl, J. E., and R. K. Herman, 1979. Polyploids and sex determination in Caenorhabditis elegans. Genetics 93: 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonel, P. E., J. Jans, B. K. Peterson and B. J. Meyer, 2006. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature 444: 614–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim, K. S., A. M. Howell and A. M. Rose, 1988. The effects of translocations on recombination frequency in Caenorhabditis elegans. Genetics 120: 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, B. J., 2005. X-chromosome dosage compensation, in WormBook, edited by The C. elegans Research Community. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Miller, L. M., J. D. Plenefisch, L. P. Casson and B. J. Meyer, 1988. xol-1: a gene that controls the male modes of both sex determination and X chromosome dosage compensation in C. elegans. Cell 55: 167–183. [DOI] [PubMed] [Google Scholar]
- Nicoll, M., C. C. Akerib and B. J. Meyer, 1997. X-chromosome-counting mechanisms that determine nematode sex. Nature 388: 200–204. [DOI] [PubMed] [Google Scholar]
- Nonet, M., and B. J. Meyer, 1991. Early aspects of Caenorhabditis elegans sex determination and dosage compensation are regulated by a zinc-finger protein. Nature 351: 65–68. [DOI] [PubMed] [Google Scholar]
- Nusbaum, C., and B. J. Meyer, 1989. The Caenorhabditis elegans gene sdc-2 controls sex determination and dosage compensation in XX animals. Genetics 122: 579–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi, M., and H. Lin, 2000. Translational repression: a duet of Nanos and Pumilio. Curr. Biol. 10: R81–R83. [DOI] [PubMed] [Google Scholar]
- Pierreux, C. E., J. Stafford, D. Demonte, D. K. Scott, J. Vandenhaute et al., 1999. Antiglucocorticoid activity of hepatocyte nuclear factor-6. Proc. Natl. Acad. Sci. USA 96: 8961–8966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plenefisch, J. D., L. Delong and B. J. Meyer, 1989. Genes that implement the hermaphrodite mode of dosage compensation in Caenorhabditis elegans. Genetics 121: 57–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, J., M. Jow and B. Meyer, 2005. The T-box transcription factor SEA-1 is an autosomal element of the X:A signal that determines C. elegans sex. Dev. Cell 9: 339–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne, M., 2004. A Genetic Switch. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Ptashne, M., and A. Gann, 2002. Genes & Signals. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Rausa, F. M., Y. Tan and R. H. Costa, 2003. Association between hepatocyte nuclear factor 6 (HNF-6) and FoxA2 DNA binding domains stimulates FoxA2 transcriptional activity but inhibits HNF-6 DNA binding. Mol. Cell. Biol. 23: 437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind, N. R., L. M. Miller, J. B. Kopczynski and B. J. Meyer, 1995. xol-1 acts as an early switch in the C. elegans male/hermaphrodite decision. Cell 80: 71–82. [DOI] [PubMed] [Google Scholar]
- Riddle, D. L., T. Blumenthal, B. J. Meyer and J. R. Priess, 1997. C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Van Gilst, M. R., H. Hadjivassiliou, A. Jolly and K. R. Yamamoto, 2005. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol. 3: e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve, A. M., and B. J. Meyer, 1987. sdc-1: a link between sex determination and dosage compensation in C. elegans. Cell 48: 25–37. [DOI] [PubMed] [Google Scholar]
- Villeneuve, A. M., and B. J. Meyer, 1990. The role of sdc-1 in the sex determination and dosage compensation decisions in Caenorhabditis elegans. Genetics 124: 91–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonker, S. A., and B. J. Meyer, 2003. Recruitment of C. elegans dosage compensation proteins for gene-specific versus chromosome-wide repression. Development 130: 6519–6532. [DOI] [PubMed] [Google Scholar]