Abstract
A gene located within the intron of a larger gene is an uncommon arrangement in any species. Few of these nested gene arrangements have been explored from an evolutionary perspective. Here we report a phylogenetic analysis of kayak (kay) and fos intron gene (fig), a divergently transcribed gene located in a kay intron, utilizing 12 Drosophila species. The evolutionary relationship between these genes is of interest because kay is the homolog of the proto-oncogene c-fos whose function is modulated by serine/threonine phosphorylation and fig is a predicted PP2C phosphatase specific for serine/threonine residues. We found that, despite an extraordinary level of diversification in the intron–exon structure of kay (11 inversions and six independent exon losses), the nested arrangement of kay and fig is conserved in all species. A genomewide analysis of protein-coding nested gene pairs revealed that ∼20% of nested pairs in D. melanogaster are also nested in D. pseudoobscura and D. virilis. A phylogenetic examination of fig revealed that there are three subfamilies of PP2C phosphatases in all 12 species of Drosophila. Overall, our phylogenetic and genomewide analyses suggest that the nested arrangement of kay and fig may be due to a functional relationship between them.
THE vast majority of genes are not nested in the introns of other genes. The first nested gene to be described in Drosophila melanogaster was located within the Gart locus (Henikoff et al. 1986). Subsequently, a set of three nested genes was identified in the dunce locus (Furia et al. 1990). In both cases, no functional relationship was identified between the nested genes. Neufeld et al. (1991) conducted the first phylogenetic analysis of a D. melanogaster nested gene pair and determined that sina and its intronic gene Rh4 were not nested in D. virilis. However, 7% of D. melanogaster genes are predicted to contain a nested gene (Adams et al. 2000), and 85% of these have a protein-coding intronic gene (15% have a noncoding RNA; Misra et al. 2002). To date, little evidence is available upon which to determine if nesting indicates a functional relationship between the genes.
Here we report a phylogenetic analysis of kayak (kay) and fos intron gene (fig), a divergently transcribed gene located in a kay intron, utilizing 12 Drosophila species. The structure and transcriptional activity of the D. melanogaster kay gene, the homolog of the human proto-oncogene c-fos, is complex and has not been fully determined. In humans, c-fos encodes part of the AP-1 transcription factor and is known to be misregulated in a number of tumors (Perkins et al. 1988). Utilizing genome annotations for D. melanogaster and confirmation with a variety of mRNA-based techniques, we generated a new model for the structure of kay (Hudson 2006). That study showed that kay is a substantial gene (27.5 kb) with three distinct promoters. In addition, nested within a large (17.5 kb) intron of kay there is a predicted, divergently transcribed gene (CG7615) that we have named fos intron gene (fig).
Our new model determined that kay has three transcription initiation sites that create alternative 5′ exons, each containing their own predicted initiator methionine (Figure 1A). Each of these 5′ exons splices to a common 3′ exon (kay-mainbody) that encodes the Basic domain (DNA binding) and the leucine-zipper domain (dimerization) essential for Kay activity. The centromere proximal promoter (most distant from kay-mainbody) gives rise to the kay-α transcript. The middle promoter generates the kay-β transcript. The closest promoter leads to the kay-γ transcript. Analysis of the divergently transcribed, nested locus fig showed that it generates an intronless transcript and encodes a protein phosphatase 2C (PP2C).
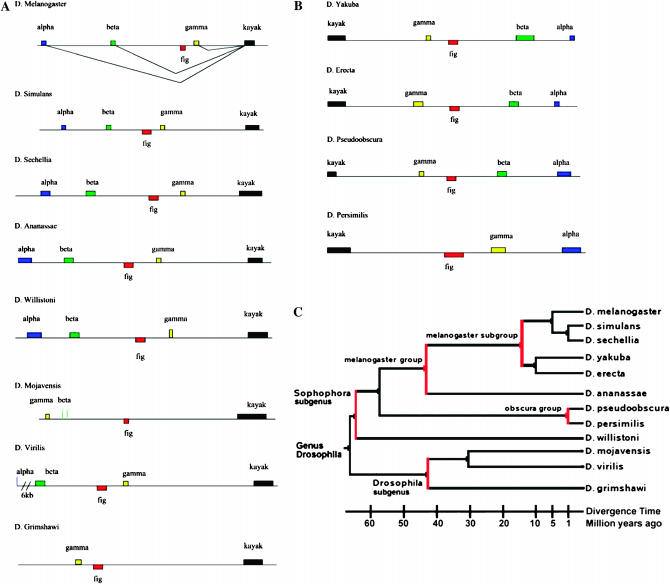
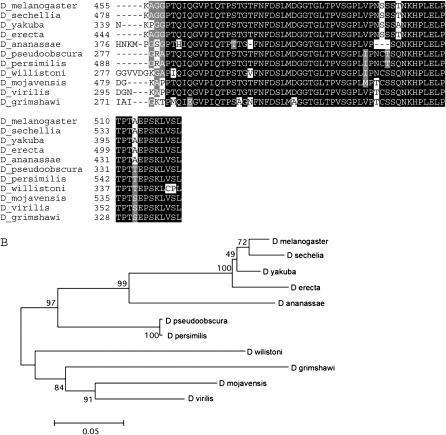
Figure 1.—
Gene structure of the kay–fig region in 12 Drosophila species. The region is shown to scale in all 12 species. The coding portions of each exon are shown in color: kay-α (blue), kay-β (green), fig (red, divergently transcribed), kay-γ (yellow), and kay-mainbody (kayak, black). (A) Eight species have the 5′-end of the kay transcription unit at the proximal end of this 27.5-kb region (closest to the centromere and depicted with 5′ to the left). The vertical blue line in D. virilis represents a segment displaying a low level of DNA sequence similarity to kay-α but no similarity at the protein level. The pair of vertical green lines in D. mojavensis represents a segment displaying a low level of DNA sequence similarity to kay-β but no similarity at the protein level. (B) Four species have a large inversion that includes this region and thus the 5′-end of the kay transcription unit is at the distal end (depicted with 5′ to the right). (C) Phylogenetic tree for the 12 Drosophila species utilized in this analysis, modified from FlyBase (2006) to match the timeline of Tamura et al. (2004).
The complexity of this region surprised us, and we wondered if each of the alternative first exons for kay and fig were conserved in distant Drosophila species. Further, we wondered if the nested arrangement of kay and fig in D. melanogaster was due to a functional relationship or was just a random recent occurrence. Numerous studies have shown that, when comparing D. virilis (subgenus Drosophila) and D. melanogaster (subgenus Sophophora), sequence conservation strongly indicates functional importance (e.g., Newfeld et al. 1993). For comparison, the 63-MY divergence between these species (Tamura et al. 2004) is roughly two-thirds of the divergence between human and mouse (93 MY; Kumar and Hedges 1998).
Evidence of a functional relationship between these genes is of interest because constitutive c-fos activity can lead to tumors and c-fos activity is stimulated by serine/threonine phosphorylation (Deng and Karin 1994). Upon activation by serine/threonine kinases, kay functions in Drosophila in the same manner as c-fos (e.g., Xia and Goldstein 1999; Ciapponi et al. 2001). How kay serine/threonine phosporylation is regulated is not thoroughly known, but the puckered serine/threonine phosphatase regulates kay activity in embryos and adults (Dobens et al. 2001). Since fig is a predicted PP2C phosphatase (specific for serine/threonine), it would not be surprising if fig plays a role in regulating kay function.
To address these questions, we examined the kay–fig genomic region in 12 species of Drosophila. We found a wide variety of gene structures for kay, as shown by the presence of multiple inversions and the repeated loss of individual kay 5′ exons. Nevertheless fig is divergently transcribed and nested in a kay intron in all species—a level of conservation that may indicate a functional relationship between them. This hypothesis is supported by our genomewide analysis of nested gene pairs that revealed that ∼20% of nested pairs in D. melanogaster are also nested in D. pseudoobscura and D. virilis. Overall, our study illustrates the power of phylogenetics to suggest experimentally testable hypotheses for the function of poorly characterized genes.
MATERIALS AND METHODS
DNA sequence retrieval:
DNA sequences were obtained at http://rana.lbl.gov/drosophila, http://evoprinter.ninds.nih.gov, and http://www.flybase.org and are attributed to the following labs: Agencourt (D. erecta, D. ananassae, D. mojavensis, D. virilis, and D. grimshawi), Washington University (D. simulans and D. persimilis), The Broad Institute (D. sechellia and D. persimilis), and Baylor University (D. pseudoobscura). Sequences corresponding to D. melanogaster accession numbers for kayak-α (DQ858474), kayak-β (AF332657, AF332658, AF332659, and AF332660; Rousseau and Goldstein 2001), kayak-γ (DQ858476), and fig (DQ858472) were utilized in BLAST to acquire homologous sequences from each of the 12 species. The FEX gene-finding program was utilized to complete predicted coding sequences as necessary (http://www.softberry.com; Solvyev and Salamov 1997). Individual sequence identifiers are listed in supplemental Tables 1–7 at http://www.genetics.org/supplemental/. Anopheles gambiae and Apis mellifera were accessed via http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi?organism=insects.
DNA sequence analysis:
Alignments of predicted amino acid sequences were generated in ClustalX (Thompson et al. 1997) and amino acid conservation highlighted with Boxshade (http://www.ch.embnet.org/software/BOX_form.html). Phylogenetic trees were generated using the neighbor-joining method with bootstrap resampling in MEGA version 3.1 (Kumar et al. 2004). Protein domains were identified via the EMBL-EBI database at http://www.ebi.ac.uk/interpro.
Genomewide studies:
For the annotation analysis, the complete set of nested protein-coding genes in D. melanogaster (Release 5.1) and the same set in D. pseudoobscura (Release 2.0) were obtained using GFF files from http://www.flybase.org. Only nested gene pairs that mimic the kay–fig structure were retrieved: two protein-coding genes with one gene completely contained within the limits of the other gene. We excluded partially overlapping genes (only an exon of one gene is within the limits of the other gene), but did include gene pairs nested on the same strand (unlike kay–fig) and on the opposite strand (like kay–fig) for completeness. We then compared these sets to identify loci where gene 1 is nested in gene 2 in D. melanogaster and the homolog of gene1 is nested in the homolog of gene 2 in D. pseudoobscura. Attributions of homology between genes in D. melanogaster and D. pseudoobscura were derived from FlyBase annotations. For the tBLASTn analysis, we began with the complete set of nested protein-coding genes in D. melanogaster (Release 5.1) obtained above. Then all exons of each nested gene pair were identified and their translated in-frame amino acid sequences were extracted. These amino acid sequences were then aligned against the D. pseudoobscura and D. virilis genomes using the tblastn algorithm (Altschul et al. 1997). tblastn results were filtered to ensure that D. pseudoobscura and D. virilis sequences matching exons of a D. melanogaster gene were located nearby on the same scaffold. Subsequently, for each nested pair in D. melanogaster, the D. pseudoobscura and D. virilis exon matches were examined to determine if matching sequence for the nested gene was located fully within the bounds of the matching sequence for the containing gene.
RESULTS
Nested arrangement of kay and fig is maintained in all 12 Drosophila species:
Our first task was to visualize the entire 27-kb kay–fig region, located on chromosome 3R at polytene band 99B10-C1 in D. melanogaster, in each of the 12 species of Drosophila that have been fully sequenced. To accomplish this, we utilized BLAST to identify and retrieve sequences corresponding to the protein-coding domain of each D. melanogaster kay exon and of fig. We found that the kay-γ, fig, and kay-mainbody coding regions are present in all species and that their location in each species fits with the chromosomal synteny identified by Muller. Each kay–fig region is located in the E group of the Muller synteny table (FlyBase 2006). This suggests that these genes were present in the common ancestor of the 12 Drosophila species.
We then determined that scaffolds surrounding each exon-specific sequence were contiguous and that fig was divergently transcribed and nested in a kay intron in all species (Figure 1). This highly conserved relationship stands out in stark contrast to the extensive diversity of gene structures for kay present in the 12 species. From our analysis, it is clear that there have been multiple chromosomal inversions and repeated loss of individual kay 5′ exons. The largest and most obvious difference among these species is a reversal in proximal–distal orientation affecting the entire kay–fig region. Eight of the species, including D. melanogaster, have the 5′-end of kay at the proximal end of the region (closest to the centromere; Figure 1A). Alternatively, four species have an inversion that includes this region and places the 5′-end of kay at the distal end of the region (closest to the telomere; Figure 1B). However, the four species with the 5′ distal arrangement are not monophyletic (Figure 1C). Therefore, the most parsimonious explanation of this distribution requires two independent inversions of the ancestral 5′ proximal orientation within the subgenus Sophophora. One inversion occurred in the branch leading to the obscura group and a second in the branch leading to D. yakuba and D. erecta in the melanogaster subgroup. As both inversions affect the entire kay–fig region, the relationship of the two sets of inversion breakpoints to each other is unknown.
In addition, multiple inversions are evident within the kay–fig region. In D. melanogaster, the relative order of the coding regions from 5′ to 3′ for kay (regardless of orientation to the centromere) is kay-α, kay-β, fig (transcribed from the opposite strand), kay-γ, and kay-mainbody. This organization is present in 9 of the 12 species. It is not present in three species where the exon order is inverted: D. persimilis, D. mojavensis, and D. grimshawi. In these three species, fig is closer to kay-mainbody than to kay-γ. However, the three species with an inversion affecting fig and kay-γ are not monophyletic (Figure 1C). As above, the most parsimonious explanation of this distribution requires three independent inversions.
There are two scenarios in which these three events could have occurred. Both scenarios require an inversion in the recent past in the D. persimilis lineage after its divergence from D. pseudoobscura. One scenario has two additional independent inversions: one in the D. grimshawi lineage and one in the D. mojavensis lineage after separation from D. virilis. Alternatively, there could have been an inversion in the subgenus Drosophila lineage leading to D. mojavensis, D. grimshawi, and D. virilis and a reversion (likely not identical to the initial inversion but one moving kay-γ and fig back to their original orientation) in D. virilis.
One consequence of the inversions that move fig closer to kay-mainbody than kay-γ is the reorientation of the fig and kay-γ open reading frames. The inversions would reverse the direction of the reading frames for fig and kay-γ in comparison to the other nine species and to their present orientation in these species. To reorient the reading frames, two small independent inversions (one each for fig and kay-γ) must be invoked in each species (six inversions total).
Evidence for these reorienting small inversions is found in D. mojavensis. Here an additional inversion involving kay-β and kay-γ reversed their order (kay-β is now closer to kay-mainbody than kay-γ; Figure 1A). This inversion rectifies the orientation of kay-γ but reverses the orientation of kay-β. The reversal of kay-β orientation suggests why the kay-β open reading frame was lost in this species. The kay-β reading frame is also absent in D. persimilis and D. grimshawi, the other two species with the inversion that moves fig closer to kay-mainbody than kay-γ, perhaps for the same reason.
If we employ chromosomal inversions as the sole mechanism for generating the diversity of gene structures seen in all species for the kay–fig genomic region, then 11 independent intragenic inversions are required. Utilizing the formula of Bartolomé and Charlesworth (2006), this equals a rate of 0.899 inversions/Mb/MY [(11 inversions/0.0275-Mb region)/445 MY total distance between all 12 species]. Other chromosomal mechanisms (e.g., reading frame maintaining transpositions) may have been involved reducing the number of events, but incorporating them would be pure speculation.
We then determined how the intragenic inversion frequency of the kay–fig region compares to published intergenic inversion frequencies. Bartolomé and Charlesworth (2006) report an intergenic inversion frequency for a two-species comparison (D. melanogaster and D. pseudoobscura) of the E group of the Muller synteny table (this includes the kay–fig region) of 0.013/Mb/MY. The rate at which we detected intragenic inversions in the kay–fig genomic region was 69-fold greater than this intergenic rate. Thus, either the kay–fig region has an anomalously high rate of inversions or the intergenic inversion frequency significantly underestimates the actual rate of inversion. Analysis of additional genes across the 12 Drosophila genomes will be needed to distinguish between these alternatives.
Genomewide analysis of nested gene pairs reveals that ∼20% maintain this arrangement in distant Drosophila species:
The absolute conservation of the nested arrangement of kay and fig contrasted sharply with the multiple inversions that we noted in the region. This led us to wonder if the conservation of nesting across distant Drosophila species for pairs of protein-coding genes was common. If it is common, then this suggests that the nested arrangement is not maintained by natural selection but rather the frequency of mutation is simply insufficient to displace what is actually a serendipitous structure. In this case, a finding of conserved nesting would indicate that the probability that the two genes are functionally related is low—on par with the likelihood that two adjacent genes have a meaningful connection.
Alternatively the conservation of nesting across distant Drosophila species for pairs of protein-coding genes could be uncommon. If it is uncommon, then this suggests that the arrangement was maintained by selection, perhaps to facilitate a functional relationship between the genes. As we could find no genomewide information on the extent of conservation for nested protein-coding gene pairs in Drosophila, we conducted this analysis by two different methods, utilizing three distantly related Drosophila species.
First, we employed a method that relied on genome annotations. Utilizing the annotations, we identified a set of 1261 nested protein-coding gene pairs in the newest release (5.1) of the D. melanogaster genome (supplemental Table 9 at http://www.genetics.org/supplemental/). Given the D. melanogaster total gene count of 14,124 (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=drosoph), these nested protein-coding genes account for ∼8.95%, a slight increase from the assessment of Adams et al. (2000). We utilized annotations to identify a set of 1363 nested protein-coding gene pairs in the latest release (2.0) of the D. pseudoobscura genome (supplemental Table 10 at http://www.genetics.org/supplemental/). A comparison of the two sets determined that 215 pairs (17.1% of those found in D. melanogaster) are nested in both species (supplemental Table 11 at http://www.genetics.org/supplemental/).
Second, we employed a method that exploited genome sequences. We extracted and translated the exons of each nested gene pair in D. melanogaster (derived from annotations as described above). These amino acid sequences were utilized to identify counterpart exons in the D. pseudoobscura and D. virilis genome sequences using tblastn. Finally, the D. pseudoobscura and D. virilis exon matches were examined to determine if a nested arrangement that corresponded to the arrangement in D. melanogaster was present. This method revealed that 391 nested pairs are conserved in D. pseudoobscura (31.0%), 376 are conserved in D. virilis (29.8%), and 318 are nested in all three species (25.2%; supplemental Table 12 at http://www.genetics.org/supplemental/).
The high degree of conservation of nesting observed in a relatively small set of genes (∼20% if one averages the results of the two analyses) supports the hypothesis that conservation may be linked to a functional relationship between the nested genes, although this is not by itself significant enough to make that argument without further evidence. Given the additional information that serine/threonine phosphatase activity is known to regulate kay and that fig is a predicted PP2C phosphatase specific for serine/threonine, we believe that the conservation of the nested relationship between these two genes in 12 Drosophila species supports the possibility that there is a functional relationship between them.
Phylogenetic analysis of kay protein-coding regions:
The coding region of the kay-α exon is present only in the nine members of the subgenus Sophophora (Figure 2A). However, in the subgenus Drosophila, D. virilis has a segment with low-level DNA similarity to the kay-α exon, but it does not encode a recognizable protein. This suggests that this species originally had the kay-α coding region, but that it has become degraded by mutation over time. Given the complete absence of the kay-α exon in the other two species in the subgenus Drosophila and that D. virilis and D. mojavensis are more closely related than D. mojavensis and D. grimshawi, we can construct two possible evolutionary histories for this exon in the subgenus Drosophila. One possibility is that the kay-α exon was lost independently and at different times in each lineage. For example, a point mutation resulting in loss of the initiator methionine would eliminate the possibility of selection maintaining the encoded protein. Alternatively, one could postulate a single loss in the lineage leading to all three subgenus Drosophila species with the caveat that the exon degraded at different rates in each lineage. We prefer the first possibility because there is no evidence of differential mutation rates in these lineages.
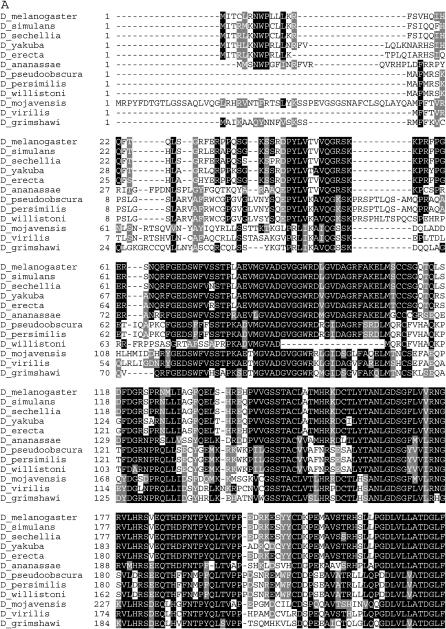
Figure 2.—
Alignment and phylogenetic tree of the kay-α exon. (A) Alignment of the coding region from nine species that have an α exon, utilizing the one-letter amino acid code. The total length of the alignment is 286 amino acids. Amino acid numbers are shown to the left. Amino acids are noted if, at a particular position, more than half of the species have the same amino acid (solid background) or a similar amino acid (shaded background). Dashes represent gaps inserted to maximize amino acid similarity between the sequences. The coordinates for each sequence are shown in supplemental Table 1 at http://www.genetics.org/supplemental/. (B) Phylogenetic tree of the coding region from nine species that have the kay-α exon. The tree is unrooted and branch lengths are drawn to scale. The scale bar shows the number of amino acid substitutions per site between two sequences. Branches with the highest bootstrap values are indicated and those >70 are considered statistically significant (Sitnikova 1996). Here, five of the eight branches are significant.
Bioinformatic analyses of this coding region show that 16% (44/286) of the positions contain an identical/similar amino acid in all species. The kay-α tree has one difference from the species tree: D. ananassae is shown as an outlier to the melanogaster group (Figure 2B). However, bootstrap support for this arrangement is weak and thus this discrepancy is not likely meaningful. Overall, our analysis suggests that the kay-α exon predates the divergence of these species, is moderately conserved, and was lost independently in three species.
The coding region of the kay-β exon is present in 9 of the 12 species (Figure 3A) and in members of both subgenera. It is not present in the three species, D. persimilis, D. mojavensis, and D. grimshawi, with an inversion affecting fig and kay-γ. In D. mojavensis, there is a segment with low-level DNA similarity to the kay-β exon, but it does not encode a recognizable protein. As discussed above, this suggests to us that D. mojavensis originally had the kay-β coding region but that it has been rendered unusable by the small inversion that rectified the orientation of the kay-γ exon. D. grimshawi and D. persimilis have no trace of the kay-β exon.
Figure 3.—
Alignment and phylogenetic tree of the kay-β exon. (A) Alignment of the coding region from nine species that have a kay-β exon. The total length of the alignment is 74 amino acids. Amino acid numbering and shading are as in Figure 2. The coordinates for each sequence are shown in supplemental Table 2 at http://www.genetics.org/supplemental/. (B) Phylogenetic tree of the coding region from nine species that have a kay-β exon. The tree is drawn as in Figure 2. Bootstrap values indicate that only three of the eight branches are significant.
Bioinformatic analyses of this coding region show that 11% (8/74) of the positions contain an identical/similar amino acid in all species. The kay-β tree has two strongly supported differences from the species tree (Figure 3B). First is the clustering of D. pseudoobscura with D. willistoni. Most likely this reflects the loss of this exon in D. persimilis. Second is the movement of D. virilis (subgenus Drosophila) between two subgenus Sophophora groups: D. ananassae (melanogaster group) and D. pseudoobscura (obscura group). Most likely this reflects the loss of this exon in the other subgenus Drosophila species. Overall, our analysis suggests that the kay-β exon predates the divergence of these species, is moderately conserved, and was lost independently in three species.
The kay-γ exon is the only one of the alternative 5′ exons that is present in all species (Figure 4A). Bioinformatic analyses of this coding region show that 74% (20/27) of the positions contain an identical/similar amino acid in all species. The kay-γ tree shows considerable differences from the species tree. However, the very short length of the alignment leads to a lack of statistical significance (Figure 4B). Overall, our analysis indicates that the kay-γ exon predates the divergence of these species and is very highly conserved.
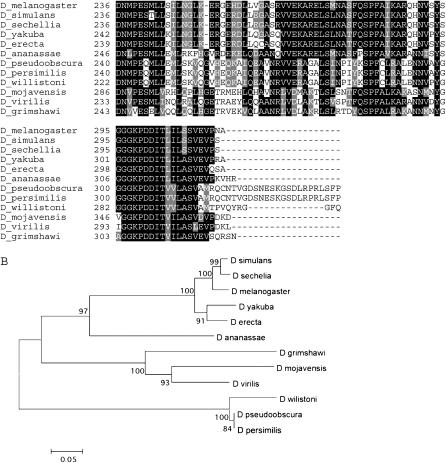
Figure 4.—
Alignment and phylogenetic tree of the kay-γ exon. (A) Alignment of the coding region from the kay-γ exon in all 12 species. The total length of the alignment is 27 amino acids. Amino acid numbering and shading are as in Figure 2. The coordinates for each sequence are shown supplemental Table 6 at http://www.genetics.org/supplemental/. (B) Phylogenetic tree of the coding region from the kay-γ exon in all 12 species. The tree is drawn as in Figure 2. Due to the short length of the alignment, bootstrap values are not significant.
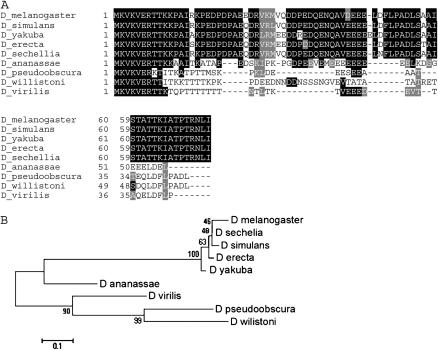
The kay-mainbody coding region is present in all 12 species (Figure 5A). We noted a gap in the middle of this sequence in D. simulans, so this species was not used in the analysis. Bioinformatic analyses found that the Basic region and leucine-zipper regions are highly conserved. Thirty-six percent (193/541) of the positions contain an identical/similar amino acid in all species. This fits well with the demonstrated functions for these two regions: the leucine zipper holds the functional c-fos dimer together and the Basic region immediately preceding the zipper binds DNA to stimulate transcription of target genes (Chen et al. 1996). The kay-mainbody tree, even without D. simulans, is identical to the species tree (Figure 5B). Overall, our analysis indicates that the kay-mainbody exon predates the divergence of these species and is well conserved.
Figure 5.—
Alignment and phylogenetic tree of the kay-mainbody exon. (A) Alignment of the coding region from the kay-mainbody exon in 11 species. What is shown begins at the equivalent of the D. melanogaster amino acid 125 because the location of the splice donor for this exon is highly variable. D. simulans was not used due to incomplete sequence in the database. The Basic region and leucine zipper begin at positions 186 and 212, respectively, in D. melanogaster. The five invariant leucines are indicated by asterisks above the D. melanogaster sequence. The total length of the alignment is 541 amino acids. Amino acid numbering and shading are as in Figure 2. The coordinates for each sequence are shown in supplemental Table 7 at http://www.genetics.org/supplemental/. (B) Phylogenetic tree of the coding region from the kay-mainbody exon in 11 species. The tree is drawn as in Figure 2. Bootstrap values indicate that 7 of the 10 branches are significant.
In summary, our analysis of kay protein-coding exons indicates that all four exons predate the divergence of these species. However, there is variation in their conservation level. The kay-α and kay-β exons are moderately conserved and have been lost independently six times. The kay-γ and kay-mainbody exons are present in all species and are highly conserved.
Phylogenetic analysis of fig:
Initially, we wondered if there were any well-characterized proteins similar to fig in other species that would suggest a function for this gene. To identify such sequences, we conducted BLASTp and protein domain searches. The domain analysis identified fig as a PP2C enzyme (InterPro database accession no. IPR000222) and, of the top 14 BLAST matches, 12 were PP2C Phosphatases (supplemental Table 8 at http://www.genetics.org/supplemental/). The other two matches (the top matches) were to other predicted D. melanogaster genes (CG15035 and CG12091). None of the BLAST matches were insubstantial as their scores ranged from 5e-61 to 5e-39 and they represented organisms from flies to humans. Thus, fig is very likely a member of a large family of PP2C phosphatases. These enzymes are serine/threonine-specific protein phosphatases that are active on a wide variety of substrates. They are found in plants, animals, and bacteria (e.g., Paramecium). Interestingly, this nearly universal species distribution is similar to that of fos (if one considers the c- and v- forms).
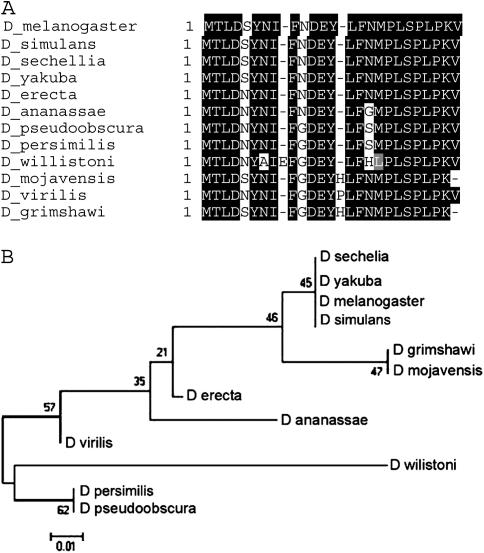
The fig coding region is present in all 12 Drosophila species (Figure 6A). Bioinformatic analyses show that fig is highly conserved. Thirty-three percent (126/382) of the positions contain an identical/similar amino acid in all species, a level of conservation nearly identical to that seen for kay-mainbody. The fig tree agrees with the species tree with two minor exceptions (Figure 6B). First, in the fig tree, D. willistoni falls within the subgenus Drosophila (with a 100% bootstrap value) instead being the most distinct species in the subgenus Sophophora. However, this is likely due to an unreadable stretch of nucleotides in the coding region that result in a gap of 19 amino acids (Figure 6A). Second, the obscura group including D. willistoni is considered more distant from the melanogaster group than the subgenus Drosophila. However, without a bootstrap value, this is likely not meaningful. Overall, our analysis indicates that fig predates the divergence of these species and is well conserved and that its evolutionary history is the same as that of kay-mainbody.
Figure 6.—
Alignment and phylogenetic tree of fig. (A) Alignment of the coding region of fig from all species. The total length of the alignment is 382 amino acids. Amino acid numbering and shading are as in Figure 2. The coordinates for each sequence are shown in supplemental Table 3 at http://www.genetics.org/supplemental/. (B) Phylogenetic tree of the coding region of fig from all species. The tree is drawn as in Figure 2. Bootstrap values indicate that 9 of the 11 branches are significant. The placement of D. willistoni in the obscura group is likely due to a gap of 19 amino acids.
We could find only a single published study of a PP2C family member in Drosophila (Dick et al. 1997). Therefore, to gather additional information on Drosophila PP2C family members, we decided to examine the relationship between fig and the two most similar genes in the database (supplemental Table 8 at http://www.genetics.org/supplemental/)—the predicted Drosophila genes CG15035 and CG12091. Given the identity of the other BLAST hits (all with less convincing matches to fig), it seems safe to assume that these are also PP2C phosphatases.
First we determined that each of these genes is present in all species. This was unsurprising as most species have multiple PP2C genes. For example, there are five PP2C genes in mammals (Jin et al. 2004). However, due to technical issues (gaps and N's) in the genome sequences, we could retrieve only 35 sequences, and three of these are not full length. D. simulans CG15035 could not be retrieved, as it is barely visible alongside a gap that takes out most of the coding region. D. ananassae CG15035 is truncated at its amino terminus, and D. persimilis CG12091 is truncated at its carboxy terminus by gaps. Also, as noted above, D. willistoni fig has an unreadable stretch of 19 amino acids. Sequence identifiers for each gene are found in supplemental Tables 3–5 (http://www.genetics.org/supplemental/).
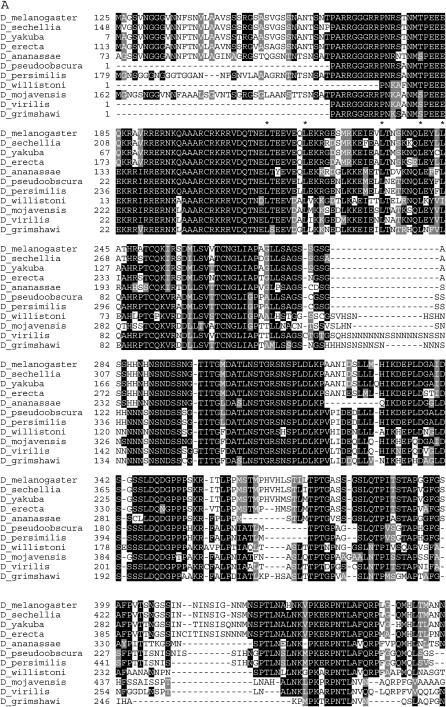
We then generated an alignment of the 35 sequences (supplemental Figure 1 at http://www.genetics.org/supplemental/). The alignment revealed that the obscura group species CG15035 genes have 5′ extensions of 130 amino acids and that their fig sequences have 3′ extensions of 19 amino acids not found in any other species. Excluding these extensions, a core stretch of ∼250 amino acids is well conserved in all sequences. Four discrete regions that encompass over half of the core sequence (∼150 amino acids) and are very highly conserved were identified. Within these regions, between 39 and 71% of the positions have an identical/similar amino acid in all species (here we ignore gaps due to the incomplete nature of several sequences).
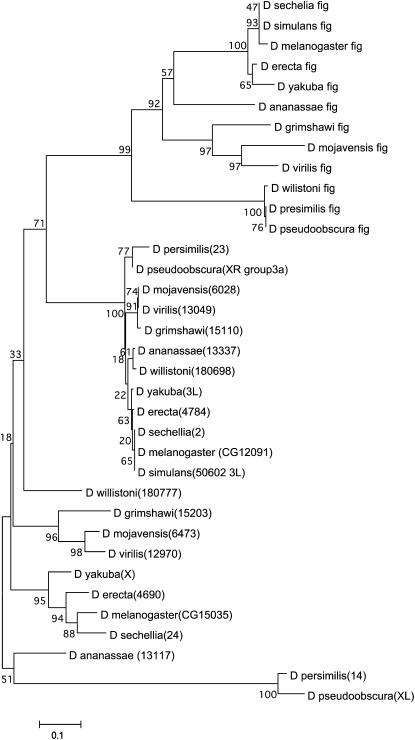
The tree of the 35 Drosophila PP2C coding regions (Figure 7) identifies three different distinct subfamilies with statistically significant support: a fig subfamily (top), a CG12091 subfamily (middle), and a CG15035 subfamily (bottom). The CG15035 sequences do not actually form a cluster like the other two in which all sequences connect to a single originating branch. The CG15035 sequences form a loose group of smaller clusters based on exclusion from the other subfamilies. Nevertheless, each subfamily contains one member from each species.
Figure 7.—
Phylogenetic tree of Drosophila PP2C phosphatases. The tree was derived from an alignment of the coding regions of 35 PP2C phosphatase sequences. See supplemental Figure 1 at http://www.genetics.org/supplemental/ for the alignment in which amino acid numbering and shading are as in Figure 2. What is shown begins at the equivalent of the D. pseudoobscura and D. persimilis CG15035 amino acid 121 as these species have unique 5′ extensions. Each of the 12 species has three PP2C phosphatases except D. simulans CG15035 could not be retrieved. D. ananassae CG15035 is truncated at its amino terminus and D. persimilis CG12091 at its carboxy terminus. D. willistoni fig has a gap of 19 amino acids. The total length of the alignment is 518 amino acids. The coordinates for each sequence are shown supplemental Tables 3–5. Numbers in parentheses identify individual sequences in supplemental Tables 4–5 and in supplemental Figure 1. Bootstrap values indicate that the presence of three subfamilies containing one sequence from each species is significant: fig (top), CG12091 (middle), and CG15035 (bottom). Within these subfamilies, CG15035 (6 of 10) and fig (8 of 11) have a majority of significant branches while CG12091 (4 of 11) has less than half of its branches as significant.
In the PP2C tree, the fig subfamily tree is identical to the tree obtained when only fig sequences were analyzed (Figure 6) and is strongly supported as 8 of its 11 branches have statistically significant bootstraps. Given the exception noted above for D. willistoni, the fig subfamily tree is different from the species tree in that it has the subgenus Drosophila between the melanogaster group and the obscura group. However, unlike the fig tree, in the PP2C tree this change has significant support.
The CG12091 subfamily tree is unaffected by the truncation of D. persimilis, and this species remains in the obscura group. The CG12091 subfamily tree is different from the species tree in two ways, one major and one minor. Like the fig subfamily tree, the CG12091subfamily tree gives statistically significant support for placing the subgenus Drosophila between the melanogaster group and the obscura group. In a minor difference, the CG12091 subfamily tree groups D. ananassae with D. willistoni. In the CG15035 subfamily tree, there are two differences from the species tree but neither is strongly supported. D. ananassae is clustered with the obscura group most likely due to its truncation and D. willistoni is again more distant from the melanogaster group than the subgenus Drosophila.
Clearly, all of the PP2C subfamilies predate the divergence of the species and each subfamily tree strongly resembles the species tree. In each subfamily tree, the subgenus Drosophila and the melanogaster subgroup (8 of the 12 species) appear as they do in the species tree. Differences between the species tree and the subfamily trees that are not explained by sequence gaps are limited to the intervening species, D. ananassae, D. willistoni, and the obscura group. One possible explanation is that when a single species is utilized to represent a large number of species (e.g., in Figure 1C, D. ananassae representing the 148 species of the melanogaster group that do not belong to the melanogaster subgroup; Ashburner 1989), stochastic changes that occurred in that lineage can have an excessive impact on that species' placement in the tree.
DISCUSSION
Our analysis provides a new illustration of the power of phylogenetic analysis to suggest experimentally testable hypotheses for the function of poorly characterized genes. We, and others, have typically employed phylogenetic analysis to address large-scale questions (e.g., Newfeld et al. 1999; Newfeld and Wisotzkey 2006), utilizing sequences from widely disparate organisms such as humans and flies that have an estimated divergence of 950 MY (Wang et al. 1999). In these studies, the goal is to determine the evolutionary relationship among members of a multigene family in different species. Information on these relationships (homology, orthology, paralogy) is then employed by experimental biologists as a framework in which to meaningfully interpret experiments outside their own model system.
The near-complete sequencing of the genomes of 12 species of Drosophila allows us to expand the application of phylogenetic analysis to address small-scale questions utilizing sequences from closely related organisms (divergence of the Sophophora and Drosophila subgenera is estimated at 63 MY; Tamura et al. 2004). Here, phylogenetics can be employed to test hypotheses about the structure of a single gene and the functional relationships among genes in the same species.
Our analysis of kay structure reveals an intriguing juxtaposition of conservation (the nested arrangement and divergent orientation of kay and fig transcription) and diversification (a 69-fold greater rate of intragenic inversion than previously reported rates of intergenic inversion). This shows, at the molecular level, the ability of natural selection to maintain the functionality of an essential gene in spite of the mutations and chromosome rearrangements that are an inevitable feature of DNA replication and mitosis/meiosis. Regarding a potential kay–fig functional relationship, our analysis reveals that fig is divergently transcribed and nested in a kay intron in all 12 species. Since our genomewide analysis determined that only 20% of the nested gene pairs in D. melanogaster are conserved in D. pseudoobscura and D. virilis, we believe that the absolute conservation of the nested relationship strongly supports the possibility that there is a functional relationship between kay and fig.
Taken together, our data also lead us to propose a model for the genetic mechanism underlying the origin and maintenance of the nested arrangement of kay and fig. For the origin, we propose a genetic event (an inversion seems likely, given their frequency in the region) predating the divergence of the 12 Drosophila species that placed fig upstream of kay in a head-to-head orientation. At that time, kay consisted of its most highly conserved exons (kay-γ and kay-mainbody) with a sole promoter region adjacent to the 5′-end of kay-γ. A head-to-head orientation of two genes requires that they be divergently transcribed, and the inversion placed the 5′-end of fig near the kay-γ promoter. This orientation still exists for the 9 species in the subgenus Sophophora. Given the proximity of the two 5′-ends, perhaps the kay-γ promoter began to influence fig transcription and it became a bidirectional promoter. Bidirectional promoters have been reported for other gene pairs with head-to-head orientations (e.g., DHFR and Rep-3 in mice; Linton et al. 1989).
In a subsequent step (or steps), also prior to the divergence of the 12 species, the kay-α and kay-β exons and their respective promoter regions were created downstream of fig with an orientation that allowed them to splice to the kay-mainbody exon. The creation of these new exons (perhaps by transposable element or illegitimate recombination mechanisms; Long 2001; Pavlicek et al. 2002) would then place fig in a kay intron. According to this model, the nested arrangement of kay and fig has been maintained because over time the kay-γ promoter became an irreplaceable component of fig transcription.
As such, any event that moved fig away from the 5′-end of kay-γ or reversed its direction of transcription would be strongly selected against or compensated for by a subsequent event. In fact, a reversal in fig transcription was not observed in any species even though fig was moved to the 3′ side of kay-γ by an inversion in three species. Evidence of a subsequent reorienting inversion affecting kay-γ is visible in one of these species and is therefore inferred in the other two. Finally, as our model has kay-α and kay-β as relatively new exons, it is not surprising that they have been lost numerous times.
We sought to gain support for this model by examining the arrangement of kay and fig in an insect outside the genus Drosophila. First, we analyzed the mosquito An. gambiae, the closest relative to Drosophila with a genome-sequencing project. Unfortunately, the kay–fig region is incomplete at this time. We determined that the kay-mainbody exon is present as two exons in mosquitoes. The first contains amino acids 173–252 and the second (located 22.8 kb downstream) contains amino acids 434–521 according to the D. melanogaster sequence (Figure 5). Upstream of the first of these exons is a gap of ∼17.7 kb. When we BLAST the mosquito genome with kay-α, kay-β, or kay-γ, we retrieve nothing meaningful possibly because they are located in the gap. A BLAST with fig retrieves only a CG12095-like sequence. Thus, at this time we obtain only negative evidence to support our model: either there is no fig in mosquitoes or perhaps it lies in the gap upstream of kay-mainbody with kay-γ. The genome of the honeybee A. mellifera is even more incomplete: no evidence of kay-mainbody (which should be present as the kay homolog c-fos is present in humans) could be obtained by BLAST.
Our model presents a number of experimentally testable hypotheses. For example, to determine if kay and fig are connected by common regulation, reporter genes can be constructed carrying the region between kay-γ and fig to learn if it contains a promoter. Further, these reporters can be designed to test our idea that this promoter is bidirectional. Alternatively, to determine if these genes have a biochemical connection, one can examine the ability of fig to dephosphorylate Kay constructs that have phosphates attached at one or more of its Jun-amino terminal kinase or ERK phosphorylation sites in cell culture assays. In addition, a comparison of gene expression patterns may indicate that fig or another of the PP2C phosphatases substantially overlaps with kay. New information gained from these experiments will immediately suggest new avenues of investigation into the human c-fos proto-oncogene.
In summary, our analysis demonstrates that phylogenetics can be profitably employed to generate testable hypotheses regarding gene structure or the relationship between two genes in the same species, an extension of current practice. The application of this approach to understanding the kay–fig nested gene pair suggested that the arrangement was functional. With the availability of 12 sequenced Drosophila genomes, studies such as this will become an increasingly important tool for researchers.
Acknowledgments
Sudhir Kumar (Arizona State University) and Robert Wisotzkey (California State University-East Bay) provided valuable comments. M.J.G. is supported by a Medical Research Council Studentship, J.W.C. and S.E.C. are supported in part by Lawrence Berkeley Directed Research and Development Program (DE-AC02-05CH11231), and S.J.N. is supported by the National Institutes of Health (CA095875 and HG002516).
References
- Adams, M., S. Celniker, R. Holt, C. Evans, J. Gocayne et al., 2000. The genome sequence of D. melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Altschul, S., T. Madden, A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M., 1989. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Bartolomé, C., and B. Charlesworth, 2006. Rates and patterns of chromosomal evolution in Drosophila pseudoobscura and D. miranda. Genetics 173: 779–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R., P. Juo, T. Curran and J. Blenis, 1996. Phosphorylation of c-Fos at the C-terminus enhances its transforming activity. Oncogene 12: 1493–1502. [PubMed] [Google Scholar]
- Ciapponi, L., D. Jackson, M. Mlodzik and D. Bohmann, 2001. Drosophila Fos mediates ERK and JNK signals via distinct phosphorylation sites. Genes Dev. 15: 1540–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, T., and M. Karin, 1994. c-Fos transcriptional activity stimulated by H-Ras-activated protein kinase distinct from JNK and ERK. Nature 371: 171–175. [DOI] [PubMed] [Google Scholar]
- Dick, T., S. Bahri and W. Chia, 1997. Drosophila DPP2C1: a novel member of the PP2C family. Gene 199: 139–143. [DOI] [PubMed] [Google Scholar]
- Dobens, L., E. Martin-Blanco, A. Martinez-Arias, F. Kafatos and L. Raftery, 2001. Drosophila puckered regulates Fos/Jun levels during follicle cell morphogenesis. Development 128: 1845–1856. [DOI] [PubMed] [Google Scholar]
- FlyBase, 2006. FlyBase: anatomical data, images and queries. Nucleic Acids Res. 34: D484–D488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furia, M., A. Filomena, D. Artiaco, E. Giordana and L. Polito, 1990. A new nested gene within the dunce genetic unit of Drosophila. Nucleic Acids Res. 18: 5837–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., M. Keene, K. Fechtel and J. Fristrom, 1986. Gene within a gene: nested Drosophila genes encode unrelated proteins on opposite strands. Cell 44: 33–42. [DOI] [PubMed] [Google Scholar]
- Hudson, S., 2006. Characterization of the D. melanogaster proto-oncogene kayak and its nested gene fos intronic gene. Ph.D. Thesis, Arizona State University, Tempe, AZ.
- Jin, F., L. Liu, J. Dai, S. Gu, X. Sun et al., 2004. Molecular cloning and characterization of a novel human PP2C cDNA (PP2C epsilon). Mol. Biol. Rep. 31: 197–202. [DOI] [PubMed] [Google Scholar]
- Kumar, S., and B. Hedges, 1998. A timescale for vertebrate evolution. Nature 392: 917–920. [DOI] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura and M. Nei, 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformatics 5: 150–163. [DOI] [PubMed] [Google Scholar]
- Linton, J., J. Yen, E. Selby, Z. Chen, J. Chinsky et al., 1989. Dual bidirectional promoters at the mouse dhfr locus: cloning and characterization of two mRNA classes of the divergently transcribed Rep-1 gene. Mol. Cell Biol. 9: 3058–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, M., 2001. Evolution of novel genes. Curr. Opin. Genet. Dev. 11: 673–680. [DOI] [PubMed] [Google Scholar]
- Misra, S., M. Crosby, C. Mungall, B. Matthews, K. Campbell et al., 2002. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol. 3: research0083.1–0083.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld, T., R. Carthew and G. Rubin, 1991. Evolution of gene position: chromosomal arrangement and sequence comparison of the D. melanogaster and D. virilis sina and Rh4 genes. Proc. Natl. Acad. Sci. USA 88: 10203–10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newfeld, S., and R. Wisotzkey, 2006. Molecular evolution of Smad proteins, pp. 15–35 in Smad Signal Transduction, edited by C.-H. Heldin and P. ten Dijke. Springer-Verlag, Berlin/Heidelberg, Germany/New York.
- Newfeld, S., A. Schmid and B. Yedvobnick, 1993. Homopolymer length variation in the Drosophila gene mastermind. J. Mol. Evol. 37: 483–495. [DOI] [PubMed] [Google Scholar]
- Newfeld, S., R. Wisotzkey and S. Kumar, 1999. Molecular evolution of a developmental pathway: phylogenetic analyses of TGFβ family ligands, receptors and Smad signal transducers. Genetics 152: 783–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlicek, A., O. Clay and G. Bernardi, 2002. Transposable elements encoding functional proteins: Pitfalls in unprocessed genomic data? FEBS Lett. 523: 252–253. [DOI] [PubMed] [Google Scholar]
- Perkins, K., G. Dailey and R. Tjian, 1988. Novel Jun- and Fos-related proteins in Drosophila are functionally homologous to enhancer factor AP-1. EMBO J. 7: 4265–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau, E., and E. Goldstein, 2001. Gene structure of the Drosophila melanogaster homolog of the human proto-oncogene fos. Gene 272: 315–322. [DOI] [PubMed] [Google Scholar]
- Sitnikova, T., 1996. Bootstrap test for phylogenetic trees. Mol. Biol. Evol. 13: 605–611. [DOI] [PubMed] [Google Scholar]
- Solvyev, V., and A. Salamov, 1997. GeneFinder computer tools for analysis of human and model organism genome sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 5: 294–302. [PubMed] [Google Scholar]
- Tamura, K., S. Subramanian and S. Kumar, 2004. Temporal patterns of Drosophila evolution revealed by mutation clocks. Mol. Biol. Evol. 21: 36–44. [DOI] [PubMed] [Google Scholar]
- Thompson, J., T. Gibson, F. Plewniak, F. Jeanmougin and D. Higgins, 1997. The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D., S. Kumar and S. Hedges, 1999. Divergence time estimates for the early history of animal phyla: the origin of plants, animals and fungi. Proc. Biol. Sci. 266: 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, X., and E. Goldstein, 1999. Response of Djun and Dfos mRNA abundance to signal transduction pathways in cultured cells of D. melanogaster. Mol. Biol. Rep. 26: 147–157. [DOI] [PubMed] [Google Scholar]