Abstract
Cytoplasmic male sterility (CMS), the maternally inherited failure to produce functional pollen, has been used in the breeding of sugar beet (Beta vulgaris ssp. vulgaris). At least three different sources of CMS can be distinguished from one another as well as from normal fertile cytoplasm by polymorphisms in their mitochondrial genomes. Here we analyzed 50 accessions of cultivated and wild beets to investigate the phylogenetic relationships among male-sterility-inducing and normal cytoplasms. The haplotypes were characterized by the nucleotide sequence of the mitochondrial cox2-cox1 spacer region and mitochondrial minisatellite loci. The results indicated that (1) a normal cytoplasm line, cv. TK81-O, was situated at the major core node of the haplotype network, and (2) the three sterilizing cytoplasms in question derived independently from the core haplotype. The evolutionary pathway was investigated by physical mapping study of the mitochondrial genome of a wild beet (B. vulgaris ssp. orientalis) accession BGRC56777 which shared the same mitochondrial haplotype with TK81-O, but was not identical to TK81-O for the RFLP profiles of mitochondrial DNA. Interestingly, three sets of inverted repeated sequences appeared to have been involved in a series of recombination events during the course of evolution between the BGRC56777 and the TK81-O mitochondrial genomes.
THE involvement of cytoplasmic factors has been reported in >150 plant species in which male sterility occurs (Laser and Lersten 1972). This type of male sterility, called cytoplasmic male sterility (CMS), is usually attributed to chimeric ORFs in the mitochondrial genome (Chase 2007). In some plant species, multiple forms of CMS are found (Fauron and Casper 1994; Schnable and Wise 1998). These forms can be distinguished by their unusual associated mitochondrial ORFs and by their response to nuclear-encoded, restorer-of-fertility (Rf) genes that suppress the male-sterile phenotype.
Several different sources of male-sterile cytoplasms have been described in cultivated beets (Beta vulgaris ssp. vulgaris) and their wild progenitor, all of which are assigned into the section Beta (syn. Vulgares) of the genus Beta (Mikami et al. 1985; Saumitou-Laprade et al. 1993). The first identified was the Owen cytoplasm, discovered in the cultivar “US-1” (Owen 1945). Up to now hybrid seed production in the sugar beet has exclusively relied on Owen CMS (Bosemark 1993). We have recently found that a mitochondrial 35-kDa protein is most likely responsible for Owen CMS (Satoh et al. 2004; Yamamoto et al. 2005). The protein proved to be the product of a 387-codon mitochondrial ORF (designated preSatp6), which is fused in frame with the downstream atp6 ORF (Yamamoto et al. 2005). Interestingly, this protein was not detectable in the anther tissue of two different sources of CMS [I-12CMS(2) and I-12CMS (3)] from wild Beta beets collected in Turkey and Pakistan; instead, a CMS-associated protein of 12 kDa was identified (Hallden et al. 1992; M. P. Yamamoto, Y. Onodera, T. Kubo and T. Mikami, unpublished results). These three male-sterile cytoplasms showed different patterns of male-sterility maintenance and male-fertility restoration when crossed with the same pollen parents (T. Kubo, T. Kinoshita and T. Mikami, unpublished data). Moreover, Ducos et al. (2001) reported that male sterility caused by the G cytoplasm of wild Beta beets was associated with a mitochondrial cox2 gene lacking eight highly conserved, C-terminal amino acids. Beta beets thus appear to maintain several distinct CMS cytoplasms, each capable of conferring male sterility by an apparently different mechanism.
In this article, we attempted to obtain information on the evolutionary relationships among these male-sterility-inducing and nonsterilizing, normal cytoplasms. For this purpose, we determined the nucleotide sequence of the cox2-cox1 intergenic spacer region from a number of cultivated and wild beets as well as CMS accession lines. In the sugar beet mitochondrial genome, the cox2 gene was shown to reside 1498 bp upstream of cox1 and no genic sequences (ORFs) were found within the intergenic spacer (Kubo et al. 2000; Satoh et al. 2004). This linkage proved to be well conserved in the mitochondrial genomes of Beta species belonging to sections Beta and Corollinae (Senda et al. 1995), making the cox2-cox1 spacer a likely site for an approximation of the neutral point-mutation rate. Furthermore, we previously identified variable number tandem repeats (VNTRs) in the mitochondrial genomes of Beta beets (Nishizawa et al. 2000). Here the VNTRs were also used to evaluate relationships among the mitochondrial genomes of Beta beets. During these analyses and the mitochondrial DNA (mtDNA) RFLP assay, we became aware of the existence of the mitochondrial genome (B. vulgaris ssp. orientalis acc. BGRC56777), which resembled that of the normal cytoplasm more than any other characterized mtDNAs. We present the physical map of this genome and a detailed comparison with the normal and Owen mtDNA physical maps.
MATERIALS AND METHODS
Plant materials and nucleic acids isolation:
Fifty accessions used in this study are listed in Table 1. TK81-MS and TK81-O are Owen CMS and its maintainer (known in sugar-beet terminology as type-O, see Bosemark 1993) sugar-beet lines, respectively. They are near isogenic except for cytotype (male sterile vs. male fertile) and their mitochondrial genomes were completely sequenced in our laboratory (Kubo et al. 2000; Satoh et al. 2004). I-12CMS(2) and I-12CMS(3) possess the male-sterile cytoplasms derived from wild beets collected in Turkey and Pakistan, respectively (Mikami et al. 1985). Total cellular DNA was isolated according to Doyle and Doyle (1990). MtDNA was isolated according to Mikami et al. (1985).
TABLE 1.
Summary of haplotypes defined by the cox2-cox1 intergenic spacer and minisatellite loci in the mitochondrial genomes of cultivated and wild beets
| Species | Accession | Sequence type | Indel type | Minisatellite | Chondriome group | Origina | Sourceb |
|---|---|---|---|---|---|---|---|
| Section Beta (syn. Vulgares) | |||||||
| B. vulgaris ssp. vulgaris | TK81-O | seq01 | ind05 | min19 | A | Japanese cultivar | NARCH |
| B. vulgaris ssp. vulgaris | TK81-MS | seq07 | ind02 | min04 | B | Japanese cultivar | NARCH |
| B. vulgaris ssp. vulgaris | I-12CMS(2) | seq08 | ind05 | min06 | A | Turkey | GW |
| B. vulgaris ssp. vulgaris | I-12CMS(3) | seq09 | ind05 | min06 | A | Pakistan | GW |
| B. vulgaris ssp. vulgaris | BGRC58265-MS | seq05 | ind02 | min04 | B | Turkey | cpro-dlo |
| B. vulgaris ssp. vulgaris | BGRC58265-MF | seq01 | ind05 | min11 | A | Turkey | cpro-dlo |
| B. vulgaris ssp. vulgaris | BGRC56743 | seq14 | ind03 | min17 | F | Greece | FAL |
| B. vulgaris ssp. vulgaris | BETA98/80 | seq10 | ind05 | min16 | A | — | IPK |
| B. vulgaris ssp. vulgaris | BETA78/90 | seq02 | ind05 | min06 | A | — | IPK |
| B. vulgaris ssp. vulgaris | BETA77/86 | seq01 | ind05 | ND | A | — | IPK |
| B. vulgaris ssp. vulgaris | BETA35/85 | seq01 | ind05 | min15 | A | Tunisia | IPK |
| B. vulgaris ssp. vulgaris | BETA31/81 | seq05 | ind02 | min04 | B | — | IPK |
| B. vulgaris ssp. vulgaris | BETA29/82 | seq01 | ind05 | ND | A | — | IPK |
| B. vulgaris ssp. vulgaris | BETA192/92 | seq01 | ind05 | ND | A | — | IPK |
| B. vulgaris ssp. vulgaris | BETA184/82 | seq01 | ind05 | ND | A | USSR | IPK |
| B. vulgaris ssp. vulgaris | BETA179/73 | seq01 | ind05 | ND | A | China | IPK |
| B. vulgaris ssp. vulgaris | 753994 | seq01 | ind05 | ND | A | — | IPK |
| B. vulgaris ssp. vulgaris | 753962 | seq13 | ind02 | min04 | B | — | IPK |
| B. vulgaris ssp. vulgaris | 753932 | seq12 | ind05 | min06 | A | — | IPK |
| B. vulgaris ssp. vulgaris | 753931 | seq11 | ind05 | min10 | A | — | IPK |
| B. vulgaris ssp. orientalis | BGRC56777 | seq01 | ind05 | min06 | A | — | FAL |
| B. vulgaris ssp. maritima | PI546530-337 | seq04 | ind05 | min12 | A | Italy | USDA |
| B. vulgaris ssp. maritima | PI518369-311 | seq01 | ind05 | min10 | A | England | USDA |
| B. vulgaris ssp. maritima | PI504186-119 | seq22 | ind05 | min06 | A | Italy | USDA |
| B. vulgaris ssp. maritima | FR4-31 | seq06 | ind05 | min01 | C | France | USDA |
| B. vulgaris ssp. maritima | BGRC57715-MS | seq24 | ind05 | min06 | A | Portugal | cpro-dlo |
| B. vulgaris ssp. maritima | BGRC57713-MS | seq02 | ind05 | min06 | A | Portugal | cpro-dlo |
| B. vulgaris ssp. maritima | BGRC57712 | seq21 | ind05 | min13 | A | Portugal | FAL |
| B. vulgaris ssp. maritima | BGRC57705-MS | seq03 | ind05 | min12 | A | — | cpro-dlo |
| B. vulgaris ssp. maritima | BGRC57705-MF | seq23 | ind05 | min12 | A | — | cpro-dlo |
| B. vulgaris ssp. maritima | BGRC56773 | seq01 | ind05 | min10 | A | Italy | FAL |
| B. vulgaris ssp. maritima | BGRC54842 | seq20 | ind04 | min18 | D | Portugal | FAL |
| B. vulgaris ssp. maritima | BGRC54841 | seq01 | ind05 | min10 | A | Ireland | FAL |
| B. vulgaris ssp. maritima | BGRC54772 | seq19 | ind05 | min07 | A | Spain | FAL |
| B. vulgaris ssp. maritima | BGRC48810-MS | seq03 | ind05 | min14 | A | Tunisia | cpro-dlo |
| B. vulgaris ssp. maritima | BGRC48810-MF | seq03 | ind05 | min14 | A | Tunisia | cpro-dlo |
| B. vulgaris ssp. maritima | BGRC36478 | seq18 | ind05 | min07 | A | Greece | FAL |
| B. vulgaris ssp. maritima | BGRC35289 | seq01 | ind05 | ND | A | Turkey | FAL |
| B. vulgaris ssp. maritima | BGRC32396 | seq01 | ind05 | min10 | A | Greece | FAL |
| B. vulgaris ssp. maritima | BGRC28901 | seq05 | ind02 | min04 | B | Italy | FAL |
| B. vulgaris ssp. maritima | BETA280/91 | seq01 | ind05 | min19 | A | — | IPK |
| B. vulgaris ssp. maritima | BETA156/86 | seq17 | ind05 | min12 | A | Belgium | IPK |
| B. vulgaris ssp. maritima | BETA140/83 | seq16 | ind05 | min02 | A | — | IPK |
| B. vulgaris ssp. maritima | BETA(29)18/82 | seq01 | ind05 | ND | A | — | IPK |
| B. vulgaris ssp. maritima | BETA(29)17/85 | seq15 | ind05 | min10 | A | France | IPK |
| B. vulgaris ssp. maritima | AM4247-295 | seq01 | ind05 | min10 | A | Spain | USDA |
| B. vulgaris ssp. maritima | AM4223-230 | seq01 | ind05 | ND | A | Greece | USDA |
| B. vulgaris ssp. macrocarpa | Canary Island | seq25 | ind05 | min05 | A | Canary Islands | NARCH |
| B. vulgaris ssp. adanensis | Egypt | seq04 | ind05 | min09 | A | Egypt | NARCH |
| Section Corollinae | |||||||
| B. trigyna | WB47 | seq26 | ind01 | min03 | F | — | NARCH |
Dashes indicate unknown
Original place of collection.
The places from which we obtained seed samples: NARCH, National Agricultural Research Center for Hokkaido Region, Japan; cpro-dlo, Centre for Plant Breeding and Reproduction Research; FAL, BA Zuchtungsforschung Genbank; USDA, USDA Agriculture Research Service, U. S. Agricultural Research Station; GW, The Great Western Sugar Company; IPK, Institüt für Pflanzengenetik und Kulturpflanzenforschung Gatersleben.
PCR and sequencing:
The primers used to amplify the cox2-cox1 intergenic spacer are 5′-AAGAAGCTTCGCTCGCTCGCTCTAACGC-3′ and 5′-CTGGATCCGCTGATAGATCTGCGCCTCG-3′ (HindIII and BamHI sites were introduced to facilitate cloning and are underlined). The sequences correspond to nucleotide 237,196–237,223 and 237,966–237,993 in TK81-O mtDNA (BA000009), respectively. The sequences of four mitochondrial tandem repeat loci (TR1, TR2, TR3, and TR4) were amplified using primers described in (Nishizawa et al. 2000). Primer sets for the amplification of B. vulgaris ssp. orientalis acc. BGRC56777 mtDNA regions were: 5′-CCTCAGATCTATGACCTCCC-3′ and 5′-CCGCAAACGAAGGCTAACCC-3′ for region 1, 5′-AGTTCTTAAAGTTCCATGCG-3′ and 5′-TAAACAACCCTTTCTCTCAC-3′ for region 3, 5′-TTCGAGAGGCGAAGAATAGC-3′ and 5′-GTACTGAGTTTCCTCCTCCC-3′ for region 4, 5′-TTCGAATCCCTCTCTTTCCGTA-3′ and 5′-TGGTACAGTAGCATCCTGCC-3′ for region 5, and 5′-TCAAAACCTTTGCGCTTCAG-3′ and 5′-GGATTAGCCGATCGCTTTCG-3′ for region 6. PCR was done according to McPherson et al. (1991). The PCR products were sequenced either directly or after cloning into the pBluescript vector (Stratagene, La Jolla, CA). The lambda clone was sequenced by shotgun-sequencing strategy (Kubo et al. 2000). The sequence was determined on both strands using ABI377 (Applied Biosystems, Foster City, CA) or Li-COR4000L (Li-COR, Lincoln, NE) DNA sequencers.
Data analysis:
Nucleotide sequences were aligned with the aid of ClustalW software at http://www.ddbj.nig.ac.jp/E-mail/homology-j.html then manually adjusted in the most parsimonious way. Ambiguous sites were excluded from analysis. A statistical parsimony network was constructed using TCS (ver. 1.21) software (Clement et al. 2000). The initial network was drawn with default parameters, and then the connection limit was increased to 55 steps to connect Seq14 (BGRC56743) and seq26 (WB47).
Construction of genomic library and physical mapping of mtDNA:
MtDNA prepared from B. vulgaris ssp. orientalis acc. BGRC56777 was partially digested with MboI and then overlaid on continuous sucrose density gradients (Sambrook et al. 1989). DNA fragments of ∼18 kbp were fractionated by centrifugation and cloned into the Lambda DASH II vector (Stratagene) according to standard procedures. All other library manipulations were as described by Kubo et al. (1995, 1999). DNA fragments were labeled using Gene Images (GE Healthcare Bio-Sciences, Piscataway, NJ). Families of overlapping phages were identified by screening the acc. BGRC56777 mtDNA library with the subfragments of TK81-O mtDNA (Kubo et al. 1995) or TK81-MS mtDNA (Kubo et al. 1999). The restriction maps of the phage clones identified were determined and further extended by genome walking from the extremities of the isolated groups of phages. In this approach, distal restriction fragments of a family of clones were used to probe the phage library of BGRC56777 from which new overlapping clones were identified and mapped.
RESULTS AND DISCUSSION
Sequence variation of the cox2-cox1 spacer region:
Total DNA was prepared from 49 accessions of the section Beta as well as from one accession of the section Corollinae, and was subjected to PCR-amplification using a set of primers targeting a part (∼800 bp) of the cox2-cox1 spacer region. The amplified DNA fragments showed a clear single-band product on an agarose gel, which was subsequently sequenced either directly or after cloning. The sequence alignment yielded 102 site mutations (nucleotide substitutions) and six indels (1–128 bp) when all accessions were considered.
Phylogeny of the cox2-cox1 spacer sequences:
Nucleotide substitutions:
The use of indels as characters in parsimony analysis is logical, but their weighting relative to nucleotide substitutions is problematic. Here we presented results from analysis only when indels were coded as missing data. Informative polymorphisms allowed us to define a total of 26 distinct haplotypes of mitochondrial cox2-cox1 spacer among 50 accessions examined (Table 1). A normal cytoplasm line, TK81-O, was found to share the same haplotype (named seq01) with 8 B. vulgaris ssp. vulgaris, 9 B. vulgaris ssp. maritima, and 1 B. vulgaris ssp. orientalis accessions. Seq01 was the most frequently observed haplotype. The number of accessions having each of the other 25 haplotypes ranged from 1 to 3 (Table 1).
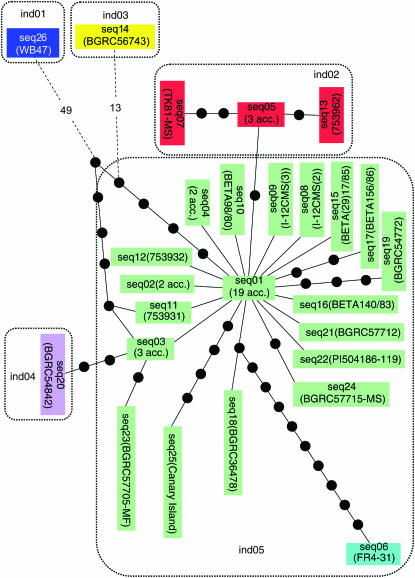
Statistical parsimony applied to our data led to the construction of the haplotype phylogeny depicted in Figure 1. The most abundant haplotype, seq01, was found to occupy the central position of the haplotype network. Eighteen terminal haplotypes (seq02, seq03, seq04, seq08, seq09, seq10, seq11, seq12, seq15, seq16, seq17, seq18, seq19, seq21, seq22, seq23, seq24, and seq25) coalesced to the internal seq01 with one to five nucleotide substitutions; phylogenetically, this group of haplotypes (provisionally named chondriome group A) in the network branched into a structure with a star-like pattern. Interestingly, two haplotypes, seq08 and seq09, were associated with male-sterile cytoplasms, I-12CMS(2) and I-12CMS(3), respectively, and our data indicate a close relationship among these two sterilizing cytoplasms and normal (“TK81-O-type”) cytoplasm.
Figure 1.—
The minimum spanning network relating 26 haplotypes defined on the basis of mitochondrial cox2-cox1 spacer sequences in cultivated and wild beets. The haplotypes are enclosed by dotted lines according to the type of indels. These 26 haplotypes were tentatively assigned into six chondriome groups: A (green), B (red), C (sky blue), D (purple), E (yellow), and F (deep blue). Haplotypes are represented by rectangles, and substitutions by black dots on the solid black lines connecting the rectangles. Dashed lines with numbers indicate the steps separating the haplotypes.
The seq01 haplotype connected to seq05, beyond which was placed haplotype seq07 (TK81-MS, Owen-sterilizing cytoplasm). The Owen CMS mtDNA haplotype was distinguished from seq05 and seq01 by three and five mutations, respectively. This indicates that the TK81-O-type normal cytoplasm is more similar to the I-12CMS(2) and I-12CMS(3) cytoplasms than to the Owen cytoplasm.
The seq05 and seq07 haplotypes formed a separate group (chondriome group B) together with seq13; this is also supported by both indel and minisatellite analyses (see below). Similarly, it might be better to consider seq06, seq20, seq14, and seq26 as four separate groups—chondriome groups C, D, E, and F, respectively. Since the outgroup seq26 (B. trigyna acc. WB47) was connected to seq01, seq11, and seq03, chondriome groups B, C, and D may be derived from chondriome group A. From seq01, three of the lineages gave rise to the Owen, I-12CMS(2) and I-12CMS(3) sterilizing mitotypes. This finding is consistent with the results of chloroplast DNA analysis (Fenart et al. 2006), which also revealed that the four different sources of beet CMS (E, G, H, and Owen types) did not constitute a single evolutionary lineage but occurred independently from an ancestral nonsterilizing cytoplasm.
Indels:
Six indels could distinguish five haplotypes (ind01–ind05), as detailed in Figure 2 and Table 2. As might have been expected, almost all indels were correlated with an outgroup, B. trigyna (ind01). Most of the accessions examined shared the ind05 haplotype, which showed four indel differences from the ind01 haplotype. The ind02 haplotype was found in the five members of chondriome group B (TK81-MS, BETA31/81, 753962, BGRC28901, and BGRC58265-MS) and differed from the three other haplotypes (ind01, ind03, and ind04) by one or two indels. The ind03 and ind04 corresponded to chondriome groups E (BGRC56743) and D (BGRC54842), respectively.
Figure 2.—
Nucleotide sequence alignment of the portions of cox2-cox1 spacer sequences from WB47 (ind01), TK81-MS (ind02), BGRC56743 (ind03), BGRC54842 (ind04), and TK81-O (ind05), representing five indel-based haplotypes. Dashes are introduced for the maximum matching.
TABLE 2.
Insertions/deletions found in the mitochondrial cox2-cox1 spacer region
| Haplotype | Species | Accession |
|---|---|---|
| ind01 | B. trigyna | WB47 |
| ind02 | B. vulgaris ssp. vulgaris | TK81-MS |
| B. vulgaris ssp. vulgaris | BETA31/81 | |
| B. vulgaris ssp. vulgaris | 753962 | |
| B. vulgaris ssp. maritima | BGRC28901 | |
| B. vulgaris ssp. vulgaris | BGRC58265-MS | |
| ind03 | B. vulgaris ssp. vulgaris | BGRC56743 |
| ind04 | B. vulgaris ssp. maritima | BGRC54842 |
| ind05 | The others | |
MtDNA minisatellite polymorphism:
Four tandem repeat loci (TR1, TR2, TR3, and TR4) were shown to be variable in the mitochondrial genomes of beets (Nishizawa et al. 2000; Fievet et al. 2007). These minisatellite sequences were used to study genetic relationships among 41 accessions representing all the haplotypes of mitochondrial cox2-cox1 spacer. All four loci analyzed were polymorphic in the material examined, with the number of alleles per locus ranging from 2 to 10 (Table 3). Heteroplasmy, or the presence of multiple-size classes of minisatellites within individual mtDNA samples, was not detected in any samples.
TABLE 3.
Number of the repeat units in the four mitochondrial minisatellite (TR) loci of cultivated and wild beets
| Haplotype | Chondriome group | Species | Accession | Sequence type | TR1 | TR2 | TR3 | TR4 |
|---|---|---|---|---|---|---|---|---|
| min01 | C | B. vulgaris ssp. maritima | FR4-31a | seq06 | 2 | 3 | 1 | 3 |
| min02 | A | B. vulgaris ssp. maritima | BETA140/83 | seq16 | 3 | 3 | 1 | 3 |
| min03 | F | B. trigyna | WB47 | seq26 | 3 | 3 | 3 | 3 |
| min04 | B | B. vulgaris ssp. vulgaris | BETA31/81 | seq05 | 4 | 3 | 2 | 4 |
| B | B. vulgaris ssp. vulgaris | BGRC58265-MS | seq05 | 4 | 3 | 2 | 4 | |
| B | B. vulgaris ssp. maritima | BGRC28901 | seq05 | 4 | 3 | 2 | 4 | |
| B | B. vulgaris ssp. vulgaris | TK81-MSa | seq07 | 4 | 3 | 2 | 4 | |
| B | B. vulgaris ssp. vulgaris | 753962 | seq13 | 4 | 3 | 2 | 4 | |
| min05 | A | B. macrocarpa | Canary Island | seq25 | 4 | 3 | 3 | 3 |
| min06 | A | B. vulgaris ssp. orientalis | BGRC56777 | seq01 | 5 | 3 | 2 | 3 |
| A | B. vulgaris ssp. vulgaris | BETA78/90 | seq02 | 5 | 3 | 2 | 3 | |
| A | B. vulgaris ssp. vulgaris | I-12CMS(2) | seq08 | 5 | 3 | 2 | 3 | |
| A | B. vulgaris ssp. vulgaris | I-12CMS(3)a | seq09 | 5 | 3 | 2 | 3 | |
| A | B. vulgaris ssp. maritima | PI504186-119 | seq22 | 5 | 3 | 2 | 3 | |
| A | B. vulgaris ssp. maritima | BGRC57715-MS | seq24 | 5 | 3 | 2 | 3 | |
| A | B. vulgaris ssp. vulgaris | 753932 | seq12 | 5 | 3 | 2 | 3 | |
| A | B. vulgaris ssp. maritima | BGRC57713-MS | seq02 | 5 | 3 | 2 | 3 | |
| min07 | A | B. vulgaris ssp. maritima | BGRC36478 | seq18 | 5 | 3 | 3 | 3 |
| A | B. vulgaris ssp. maritima | BGRC54772 | seq19 | 5 | 3 | 3 | 3 | |
| min08 | A | B. vulgaris ssp. adanensis | Egypt | seq04 | 6 | 3 | 2 | 3 |
| min09 | A | B. vulgaris ssp. maritima | AM4247-295a | seq19 | 6 | 3 | 3 | 3 |
| A | B. vulgaris ssp. maritima | BETA(29)17/85 | seq15 | 6 | 3 | 3 | 3 | |
| A | B. vulgaris ssp. vulgaris | 753931 | seq11 | 6 | 3 | 3 | 3 | |
| A | B. vulgaris ssp. maritima | PI518369-311 | seq01 | 6 | 3 | 3 | 3 | |
| A | B. vulgaris ssp. maritima | BGRC56773 | seq01 | 6 | 3 | 3 | 3 | |
| A | B. vulgaris ssp. maritima | BGRC54841 | seq01 | 6 | 3 | 3 | 3 | |
| A | B. vulgaris ssp. maritima | BGRC32396 | seq01 | 6 | 3 | 3 | 3 | |
| min10 | A | B. vulgaris ssp. vulgaris | BGRC58265-MF | seq01 | 7 | 3 | 2 | 3 |
| min11 | A | B. vulgaris ssp. maritima | BGRC57705-MS | seq03 | 7 | 3 | 3 | 3 |
| A | B. vulgaris ssp. maritima | BGRC57705-MF | seq23 | 7 | 3 | 3 | 3 | |
| A | B. vulgaris ssp. maritima | PI546530-337 | seq04 | 7 | 3 | 3 | 3 | |
| A | B. vulgaris ssp. maritima | BETA156/86 | seq17 | 7 | 3 | 3 | 3 | |
| min12 | A | B. vulgaris ssp. maritima | BGRC57712 | seq21 | 7 | 3 | 3 | 4 |
| min13 | A | B. vulgaris ssp. maritima | BGRC48810-MF | seq03 | 8 | 3 | 3 | 5 |
| A | B. vulgaris ssp. maritima | BGRC48810-MS | seq03 | 8 | 3 | 3 | 5 | |
| min14 | A | B. vulgaris ssp. vulgaris | BETA35/85 | seq01 | 8 | 6 | 3 | 1 |
| min15 | A | B. vulgaris ssp. vulgaris | BETA98/80 | seq10 | 9 | 3 | 3 | 3 |
| min16 | E | B. vulgaris ssp. vulgaris | BGRC56743 | seq14 | 9 | 3 | 2 | 5 |
| min17 | D | B. vulgaris ssp. maritima | BGRC54842 | seq20 | 11 | 3 | 3 | 3 |
| min18 | A | B. vulgaris ssp. vulgaris | TK81-Oa | seq01 | 13 | 3 | 3 | 3 |
| A | B. vulgaris ssp. maritima | BETA280/91 | seq01 | 13 | 3 | 3 | 3 |
Data are from Nishizawa et al. (2000).
Eighteen multi-locus haplotypes (min01–min18) were identified, of which 13 were observed in the accessions assigned into the chondriome group A (Tables 1 and 3). Chondriome group B was monomorphic for haplotype min04, which was not found in any other chondriome groups and this group thus proved to be clearly separated from the others. Chondriome groups C, F, E, and D were characterized by unique min01, min03, min16, and min17, respectively, none of which was found in the other chondriome groups.
A physical map of the mitochondrial genome of B. vulgaris ssp. orientalis acc. BGRC56777:
The seq01 haplotype has attracted our interest because it most likely represents the progenitor common to the Owen, I-12CMS(2) and I-12CMS(3) sterilizing mitotypes. One can expect that more intensive comparative analysis on the mitochondrial genomes of beet accessions carrying the seq01 haplotype would further subdivide this haplotype, allowing the maternal lineage of the three sterilizing mitotypes to be unambiguously traced. As a first step toward this end, we constructed a physical map of the mitochondrial genome of B. vulgaris ssp. orientalis acc. BGRC56777 (seq01 haplotype) and compared it with the other known sugar beet mtDNA maps (Kubo et al. 1995, 1999). BGRC56777 was chosen for mapping because this accession was found to be similar but not identical to cv. TK81-O for the RFLP profiles of mtDNA and was also considered to possess the fertile cytoplasm (S. Nishizawa, T. Mikami and T. Kubo, unpublished data).
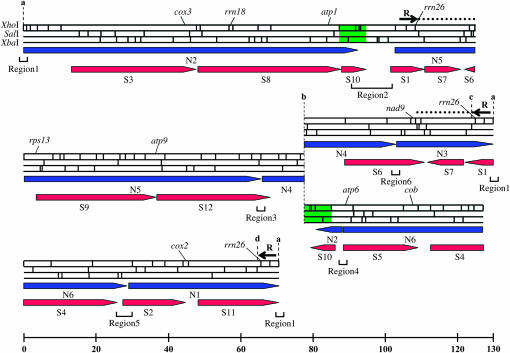
Two sets of overlapping phage clones, each spanning the entire mitochondrial genomes of cv. TK81-O and cv. TK81-MS (Kubo et al. 1995, 1999), respectively, were sequentially used as probes against Southern blots prepared with total genomic DNA from BGRC56777. This enabled us to identify a number of regions that were unchanged in hybridization patterns either between BGRC56777 and TK81-O or between BGRC56777 and TK81-MS, but that were delimited by rearrangement breakpoints. We further adopted the genome-walking technique where fragments located at the extremities of the homologous sequence regions were used to probe a BGRC56777 phage library from which overlapping clones were identified and mapped. In this manner, the entire sequence complexity was mapped onto two circular chromosomes of 257.6 and 324.3 kb, which share 210.6-kb of homologous sequence. A master circle (581.9 kb) can also be constructed with the assumption that recombination may happen via the 210.6-kb repeats (Figure 3).
Figure 3.—
A physical map of the mitochondrial genome of B. vulgaris ssp. orientalis acc. BGRC56777. Three rearrangement points (designated b, c, and d) were identified. The 47.0-kb region from b to c and the 113.7-kb region from b to d do not have features in common. The ends of these two single-copy regions can be connected to the end (top left) of the map. Black horizontal arrows labeled by the letter R indicate the active recombination repeated sequences containing rrn26. Dotted lines show the extension of the R repeat. Green-boxed sequences are repeated sequences but not recombinogenic. The regions of homology between the BGRC56777 and the TK81-O mtDNA and between the BGRC56777 and the TK81-MS mtDNAs are indicated by blue arrows designated N1–N6 and red arrows designated S1–S12, respectively. The sequenced regions (regions 1–6) are indicated by brackets.
We also found a single family of recombination-active repeats; the region common to all repeat copies was 6.1 kb in length and harbored the rrn26 gene (Figure 3). All possible combinations of flanking sequences around the repeats were detected either by restriction mapping of repeat-containing phages or by hybridizations of flanking-region probes to genomic DNA, which indicates that they are recombinogenic (data not shown).
Comparison of the mitochondrial genomes among BGRC56777, fertile line TK81-O, and Owen CMS line TK81-MS:
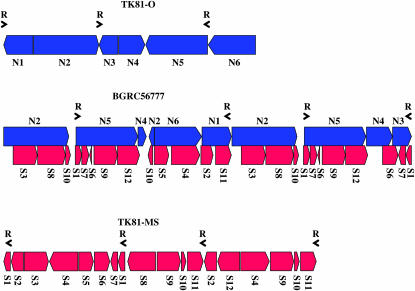
We compared the linear arrangement of the homologous restriction enzyme fragments among BGRC56777, TK81-O, and TK81-MS mtDNAs on the basis of phage clone analysis and cross-hybridization studies. The BGRC56777 mitochondrial genome could be divided into regions where boundaries or breakpoints were defined by an interruption in the restriction maps of the BGRC56777, TK81-O, or TK81-MS master chromosomes (Figure 3). When BGRC56777 was compared with TK81-O, six such regions (N1–N6) were identified in the TK81-O genome, each of which had at least one counterpart in the BGRC56777 mitochondrial genome. As shown in Figure 4, a segment of the BGRC56777 genome contains regions N2–N5 in the same order as the TK81-O genome, although the regions have different orientations. A possible evolutionary pathway for interconverting the two genomes will be presented later.
Figure 4.—
Schematic of the rearrangements among the mitochondrial genomes of BGRC56777, TK81-O, and TK81-MS. The order and relative orientation of the homologous regions (designated N1–N6 and S1–S12, see Figure 3) between the BGRC56777 and the TK81-O mtDNAs and between BGRC56777 and TK81-MS mtDNAs are shown. Black horizontal arrowheads labeled by the letter R indicate the active recombination repeated sequences. In the TK81-MS mitochondrial genome, two other recombinogenic repeated sequences (Kubo et al. 1999) are not identified.
Twelve regions (S1–S12) were delimited by comparing BGRC56777 and TK81-MS. We should note that 5 regions (S1, S2, S6, S10, and S12) spanned the organizational breakpoints between TK81-O and BGRC56777, indicating that the 5 regions are conserved between the BGRC56777 and the TK81-MS mitochondrial genomes. Figure 4 illustrates the large differences in order and orientation of the 12 regions between the two mitochondrial genomes. In addition to the rearrangements of homologous sequences, the BGRC56777 mtDNA sustained several deletions (as many as five) and insertions (as many as five) relative to the TK81-MS mtDNA. The complexity of the differences does not offer a straightforward explanation of the evolutionary relationship of the two genomes.
Rearrangements found between the mitochondrial genomes of BGRC56777 and TK81-O: events and evolutionary pathways:
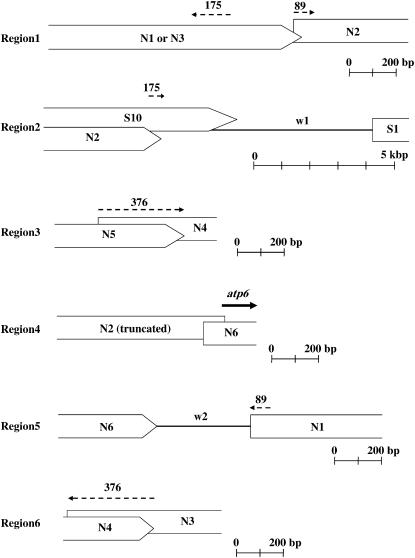
What we next describe is the possible mechanism of rearrangements detected between the mitochondrial genomes of BGRC56777 and TK81-O. The mitochondrial genomes of higher plants are characterized by an abundance of short dispersed repeats, some of which are associated with various rearrangements, including inversions, insertions/deletions, and transpositions (Fauron et al. 1995; Lilly and Havey 2001; Kubo and Mikami 2007). A complete sequence comparison of the normal and Owen CMS mtDNAs revealed in fact that most, if not all, rearrangements involved short repeats present at the recombination endpoints (Satoh et al. 2006). With this in mind, we determined the nucleotide sequences of the six regions covering the rearrangement breakpoints in the BGRC56777 mitochondrial genome and analyzed these sequences to search for repeats (see Figure 3); regions 1, 3, 4, 5, and 6 were PCR amplified, whereas the insert of a lambda phage clone (347) was used to sequence region 2 (accession numbers AB303857–AB303862). A comparison of the sequences determined here with the TK81-O mtDNA sequences successfully identified the exact boundary of regions N1–N6 (Figure 5).
Figure 5.—
Schematic of the six regions (1–6) covering the rearrangement breakpoints in the BGRC56777 mitochondrial genome (see Figure 3). Scale bars are shown under each illustration. Part of the atp6 coding region is shown by a solid arrow. Dashed arrows indicate three sets of small repeated sequences mentioned in the text. Boxes are sequences shared between the BGRC56777 and the TK81-O mitochondrial genomes or between the BGRC56777 and the TK81-MS mitochondrial genomes (see Figure 3). Solid horizontal lines between the boxes represent sequences unique to the BGRC56777 mitochondrial genome, labeled w1 and w2.
As shown in Figures 5 and 6, three sets of inverted repeats (89, 175, and 376 bp in length) were found flanking the breakpoints. Region N2 was flanked on one side by a copy of the 89-bp repeat and on the other side by a copy of the 175-bp repeat, whereas region N1 was bounded by another 89-bp repeat copy and another 175-bp repeat copy. In addition, two copies of the 376-bp repeat bracketed the region N4. These data were used to postulate the most parsimonious evolutionary pathway for interconverting the mitochondrial genomes of BGRC56777 and TK81-O.
Figure 6.—
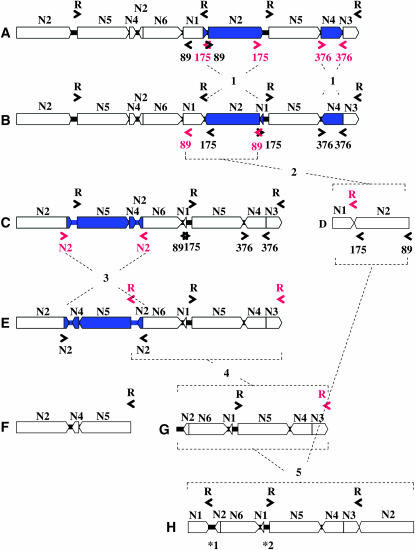
A proposed model for the evolution of the BGRC56777 mitochondrial master chromosome toward the TK81-O-like mitochondrial master chromosome. Three sets of inverted repeated sequences (89, 175, and 376 bp) were identified at the rearrangement breakpoints within the BGRC56777 mitochondrial genome (see also Figure 5), which are depicted in the linealized master chromosome A (581.9 kb). At the initial step (shown by 1), 175- and 376-bp repeated sequences (highlighted in red) recombined to introduce two inversions of the blue-colored regions, resulting in the isomeric master chromosome B. At the next step (shown by 2), 89-bp repeated sequences (highlighted in red) recombined to loop out 135.2-kb subgenomic molecule D. The remaining 446.7-kb molecule C was further converted to the isomeric molecule E by the recombination via truncated N2 repeated sequences (highlighted in red) at step 3. The molecule E was then split into two molecules, F (195.5 kb) and G (251.2 kb), by the recombination-active repeats R containing rrn26 (highlighted in red) at step 4. The molecules D and G were fused by the recombination mediated by R (highlighted in red) to yield TK81-O-like master chromosome H (386.4 kb) at step 5. *1 and *2 (parts of region 2) indicate the position of sequences unique to the 386.4-kb molecule.
The evolutionary scenario presented in Figure 6 was constructed on the basis of the assumption that the mitochondrial genome of TK81-O was derived from a BGRC56777-like ancestral genome by a succession of five major sequence rearrangements:
Recombination across the 175-bp repeat resulted in the inversion of N2, causing two copies of the 89-bp repeat to be directly oriented. The 376-bp inverted repeat mediated recombination to invert region N4.
The 89-bp direct repeat subsequently recombined to produce two subgenomic circular molecules of 135.2 and 446.7 kb, although we have no evidence bearing on the temporal order of the inversion of N4 and the generation of the 446.7-kb subgenomic molecule.
Recombination between region N2 and its truncated copy inverted the segment (128 kb) of the 446.7-kb molecule between the repeated sequences, which included one copy of the rrn26 gene and placed the two copies in direct orientation.
This was followed by recombination through rrn26, which gave rise to two subgenomic circular molecules of 195.5 and 251.2 kb.
The 135.2- and 251.2-kb subgenomic molecules underwent intermolecular recombination through the rrn26 repeat to give a molecule of 386.4 kb. The resultant master chromosome is almost identical to that of TK81-O in terms of gene order and restriction sites, but differs from the latter in having additional sequences derived from region 2 (indicated by *1 and *2 in Figure 6).
The five-step rearrangements described above seem to be the simplest and most parsimonious hypothesis to explain the evolutionary relationship of the mitochondrial genomes of BGRC56777 and TK81-O. However, we have no definite evidence supporting this progenitor (BGRC56777-like genome)/derivative (TK81-O genome) relationship. It should also be borne in mind that consideration of parsimony or simplicity may be inadequate for determining whether the proposed model may closely represent actual evolutionary events. It may be best to use comparative mapping data by examining a number of related mitochondrial genomes. We are now attempting to construct the physical maps of mitochondrial genomes of I-12CMS(2) and I-12CMS(3), as well as different sources of normal cytoplasms carrying seq01 haplotype, with the aim of gaining a better understanding of how the three sterilizing cytoplasms in question evolved.
Acknowledgments
Seeds were kindly supplied by L. Frese (Institute für Pflanzenbau der Bundesforschungsanstalt für Landwirtschaft), H. Krautkramer (Institut für Pflanzengenetik und Kulturpflanzenforschung Gatersleben), M. H. Yu (U.S. Department of Agriculture, Agriculture Research Service, U.S. Agricultural Research Station), W. Lange (Centre for Plant Breeding and Reproduction Research, The Netherlands), and Hokkaido National Agricultural Experiment Station (Japan). We gratefully acknowledge Y. Kishima, R. Masuda, S. Akimoto, and K. Yoshizawa for valuable suggestions. This work was done in part at the Research Center for Molecular Genetics, Hokkaido University, and was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, and a grant from the Program for Promotion of Basic Research Activities for Innovative Biosciences, Japan.
References
- Bosemark, N. O., 1993. Genetics and breeding, pp. 66–119 in The Sugar Beet Crops, edited by D. A. Cooke and R. K. Scott. Chapman & Hall, London.
- Chase, C. D., 2007. Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 23: 81–90. [DOI] [PubMed] [Google Scholar]
- Clement, M., D. Posada and K. A. Crandall, 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9: 1657–1660. [DOI] [PubMed] [Google Scholar]
- Doyle, J. J., and J. L. Doyle, 1990. Isolation of plant DNA from fresh tissue. Focus 12: 13–15. [Google Scholar]
- Ducos, E., P. Touzet and M. Boutry, 2001. The male sterile G cytoplasm of wild beet displays modified mitochondrial respiratory complexes. Plant J. 26: 171–180. [DOI] [PubMed] [Google Scholar]
- Fauron, C. M.-R., and M. Casper, 1994. A second type of normal maize mitochondrial genome: an evolutionary link. Genetics 137: 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauron, C. M.-R., B. Moore and M. Casper, 1995. Maize as a model of higher plant mitochondrial genome plasticity. Plant Sci. 112: 11–32. [Google Scholar]
- Fenart, S., P. Touzet, J.-F. Arnaud and J. Cuguen, 2006. Emergence of gynodioecy in wild beet (Beta vulgaris ssp. maritima L.): a genealogical approach using chloroplastic nucleotide sequences. Proc. R. Soc. London B 273: 1391–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fievet, V., P. Touzet, J.-F. Arnaud and J. Cuguen, 2007. Spatial analysis of nuclear and cytoplasmic DNA diversity in wild sea beet (Beta vulgaris ssp. maritima) populations: Do marine currents shape the genetic structure? Mol. Ecol. 16: 1847–1864. [DOI] [PubMed] [Google Scholar]
- Hallden, C., C. Lind and I. M. Moller, 1992. Variation in mitochondrial translation products in fertile and cytoplasmic male-sterile sugar-beets. Theor. Appl. Genet. 85: 139–145. [DOI] [PubMed] [Google Scholar]
- Kubo, T., Y. Satoh, T. Muro, T. Kinoshita and T. Mikami, 1995. Physical and gene organization of mitochondrial DNA from the fertile cytoplasm of sugarbeet (Beta vulgaris L.). Curr. Genet. 28: 235–241. [DOI] [PubMed] [Google Scholar]
- Kubo, T., S. Nishizawa and T. Mikami, 1999. Alterations in organization and transcription of the mitochondrial genome of cytoplasmic male sterile sugar beet (Beta vulgaris L.). Mol. Gen. Genet. 262: 283–290. [DOI] [PubMed] [Google Scholar]
- Kubo, T., S. Nishizawa, A. Sugawara, N. Itchoda, A. Estiati et al., 2000. The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNACys(GCA). Nucleic Acids Res. 28: 2571–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, T., and T. Mikami, 2007. Organization and variation of angiosperm mitochondrial genome. Physiol. Plant. 129: 6–13. [Google Scholar]
- Laser, K. D., and N. R. Lersten, 1972. Anatomy and cytology of microsporogenesis in cytoplasmic male sterile angiosperms. Bot. Rev. 38: 425–454. [Google Scholar]
- Lilly, J. W., and M. J. Havey, 2001. Small, repetitive DNAs contribute significantly to the expanded mitochondrial genome of cucumber. Genetics 159: 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson, M. J., P. Quirke and G. R. Taylor, 1991. PCR: A Practical Approach. Oxford University Press, New York.
- Mikami, T., Y. Kishima, M. Sugiura and T. Kinoshita, 1985. Organelle genome diversity in sugar beet with normal and different sources of male sterile cytoplasms. Theor. Appl. Genet. 71: 166–171. [DOI] [PubMed] [Google Scholar]
- Nishizawa, S., T. Kubo and T. Mikami, 2000. Variable number of tandem repeat loci in the mitochondrial genomes of beets. Curr. Genet. 37: 34–38. [DOI] [PubMed] [Google Scholar]
- Owen, F. V., 1945. Cytoplasmically inherited male-sterility in sugarbeet. J. Agr. Res. 71: 423–440. [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual, Ed. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Satoh, M., T. Kubo, S. Nishizawa, A. Estiati, N. Itchoda et al., 2004. The cytoplasmic male-sterile type and normal type mitochondrial genomes of sugar beet share the same complement of genes of known function but differ in the content of expressed ORFs. Mol. Genet. Genomics 272: 247–256. [DOI] [PubMed] [Google Scholar]
- Satoh, M., T. Kubo and T. Mikami, 2006. The Owen mitochondrial genome in sugar beet (Beta vulgaris L.): possible mechanisms of extensive rearrangements and the origin of the mitotype-unique regions. Theor. Appl. Genet. 113: 477–484. [DOI] [PubMed] [Google Scholar]
- Saumitou-Laprade, P., G. J. A. Rouwendal, J. Cuguen, F. A. Krens and G. Michaelis, 1993. Different CMS sources found in Beta vulgaris ssp maritima: mitochondrial variability in wild population revealed by a rapid screening procedure. Theor. Appl. Genet. 85: 529–535. [DOI] [PubMed] [Google Scholar]
- Schnable, P. S., and R. P. Wise, 1998. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3: 175–180. [Google Scholar]
- Senda, M., Y. Onodera, T. Kinoshita and T. Mikami, 1995. Mitochondrial gene variation and phylogenetic relationships in the genus Beta. Theor. Appl. Genet. 90: 914–919. [DOI] [PubMed] [Google Scholar]
- Yamamoto, M. P., T. Kubo and T. Mikami, 2005. The 5′-leader sequence of sugar beet mitochondrial atp6 encodes a novel polypeptide that is characteristic of Owen cytoplasmic male sterility. Mol. Genet. Genomics 273: 342–349. [DOI] [PubMed] [Google Scholar]