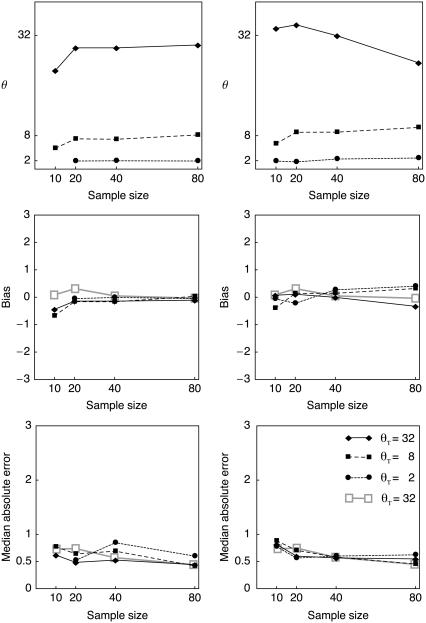
IN a recent issue of Genetics, RoyChoudhury and Stephens (2007) showcased a new method for estimating the population scaled mutation rate θ from microsatellite data; θ is equivalent to four times the effective population size times the mutation rate per generation and can also be viewed as the scaled population size. Their approximation delivered impressively accurate results with little bias. They compared their results with several other commonly available programs. Their study is a good example of how comparisons with other programs should be presented; but I was not impressed by the bias and median absolute error reported for my own program MIGRATE (Beerli and Felsenstein 2001). RoyChoudhury and Stephens (2007) used the defaults of MIGRATE and wondered, given the large observed biases, how more difficult population models would fare when MIGRATE 1.7.3 has difficulties estimating a single parameter. On my request, A. RoyChoudhury sent me their data sets, so that I could check whether the current version of MIGRATE (2.3; http://popgen.scs.fsu.edu) suffers from the same problem as the tested version. The data sets, which contained 50 unlinked microsatellite loci for sample sizes of 10, 20, 40, and 80 gene copies from a single population of size θT of 2, 8, and 32, were simulated using the coalescent simulator of Paul Fearnhead (RoyChoudhury and Stephens 2007). I ran these data sets through MIGRATE 2.3 using default settings with the stepwise mutation model and the Brownian motion approximation. A comparison of my Figure 1 with Figure 1 in their article shows clearly that the current version of MIGRATE is much less biased. In fact, the results are very similar to the approximate method of RoyChoudhury and Stephens. My Figure 1 includes their results for θT = 32 as a reference. The Brownian motion approximation in MIGRATE, already available in version 1.7.3, delivers similar results much faster; the runtime for the largest single locus data set was ∼30 sec on a 2 Ghz Opteron CPU. The microsatellite implementation in 1.7.3 seems, retrospectively, inefficient and extremely slow. The large biases were most likely a result of an aggressive default setting for a tuning parameter governing the conditional likelihood calculation and an inefficient calculation of the actual probability to make k mutational steps in time t. The effect of this tuning parameter is most pronounced with highly variable data associated with high θ values. As a result of these findings, I have changed the default for this tuning parameter. Additionally, I removed inefficiencies in the conditional likelihood calculation: this improved the runtime for the stepwise mutation model from ∼40 min on 3 Ghz machines as reported by RoyChoudhury and Stephens (2007) to ∼5 min on 2 Ghz Opteron machines.
Figure 1.—
Bias and absolute error for MIGRATE version 2.3. Each point is the median scaled mutation rate θ, bias, or error of 50 data sets per sample size and scaled population size θT. (Left) Using the stepwise mutation model; (right) using the Brownian motion approximation. Data sets, scale and calculations of bias, and absolute error are the same as in Figure 1 of RoyChoudhury and Stephens (2007); for reference, the bias and absolute error of their estimator for θT = 32, taken from their Figure 1, is shaded.
Acknowledgments
I thank Arindam RoyChoudhury and Matthew Stephens for supplying their simulated data and their explanations of their statistics and also an anonymous reviewer for helpful comments. This work was supported by the joint National Science Foundation/National Institute of General Medical Sciences mathematical biology program under National Institutes of Health grant R01 GM 078985.
References
- Beerli, P., and J. Felsenstein, 2001. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc. Natl. Acad. Sci. USA 98: 4563–4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RoyChoudhury, A., and Stephens, M., 2007. Fast and accurate estimation of the population-scaled mutation rate, θ, from microsatellite genotype data. Genetics 176: 1363–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]